Have you learned how to build a PCR-Free library?
In high-throughput sequencing, conventional library construction requires PCR amplification. On the one hand, it is to amplify trace DNA samples and increases library yield. On the other hand, it can amplify fluorescent signals, making it easier for sequencers to capture and identify fluorescent signals to improve sequencing accuracy. However, PCR is like a "double-edged sword". While solving the problem of low initial sample volume and amplifying the fluorescent signal, it also introduces amplification errors and biases, and cannot perfectly present the genome sequence "True face". In addition, PCR polymerase has a certain amplification bias. Some regions, especially high GC or low GC, secondary structure, and other regions, have low amplification efficiency and are difficult to cover. It will also introduce a large number of wrong InDels and bring a large number of Duplication. , causing a waste of DNA data volume and increasing the cost of sequencing. Therefore, the introduction of PCR-Free technology not only has the advantages of PCR but also overcomes the shortcomings of PCR. It can not only bring high-quality libraries with high sensitivity, and realize the detection of trace samples, but also have high accuracy and reduce follow-up Validation workload for variant studies. So what exactly is the PCR-Free library and what are its advantages?
1. What is PCR-Free library?
2. What are the advantages of PCR-Free technology?
3. Data display of PCR-Free
4. FAQ
5. Product information
6. Regarding reading
1. What is PCR-Free library?
Next-generation sequencing technology is the most commonly used technology in high-throughput sequencing research in modern molecular biology. Traditional first-generation sequencing can no longer fully meet the needs of researchers. Genome resequencing of model organisms and genome sequencing of non-model organisms require more cost. Low, higher throughput, faster sequencing technology, thus giving birth to the second-generation sequencing technology. The core principle of the second-generation sequencing technology is sequencing-by-synthesis, that is, to determine the DNA sequence by capturing the newly synthesized end-labeled base information. The main principle of second-generation sequencing technology for DNA sequencing is to first fragment the DNA, repair the end of the fragmented DNA, and then add specific adapters on both sides and then use different methods to generate millions of spatially fixed PCRs. For clonal arrays, sequencing data are obtained using primer hybridization and enzymatic extension reactions to image the fluorescent labels incorporated by each extension reaction.
DNA sequencing by next-generation sequencing mainly includes two processes: library preparation and on-machine sequencing. During the library preparation, randomly interrupted genomic fragments are typically amplified by standard PCR. However, for some special templates, there are factors such as complex secondary structure or poor thermal stability, which affect the amplification preference of template PCR, so not all genomic sequences can be equally reflected in the PCR amplification library. Especially for some templates with high GC or high AT content, sometimes it is difficult to use the PCR method to amplify for library construction. There is no difference between the PCR-free library and the conventional PCR library when sequencing on the machine, except that PCR is not performed during the library construction process. The PCR-free library can theoretically improve the distribution of data reads and generate more uniform genome coverage. The PCR-free library is a supplement to the library construction method. Since it is directly prepared into an onboard library without PCR amplification, it can improve the coverage of some high GC or high AT regions, thereby reducing the error bases introduced by PCR, Data bias, and sequence repetition.
The core steps of conventional NGS library construction are genome fragmentation, end repair, adapter addition, PCR, and signal amplification before sequencing. So, what about PCR-Free library construction? As the name suggests, it is a library-building process that does not require PCR. The core steps of library construction are genome fragmentation, end modification, linker addition, and library quality control. But in fact, this can only be regarded as PCR-Free in the library construction process. The real PCR-free should be a process from library construction to sequencing without library amplification (such as single-molecule sequencing technology) or a sequencing technology without accumulation of PCR amplification errors (such as MGI's DNBseq).
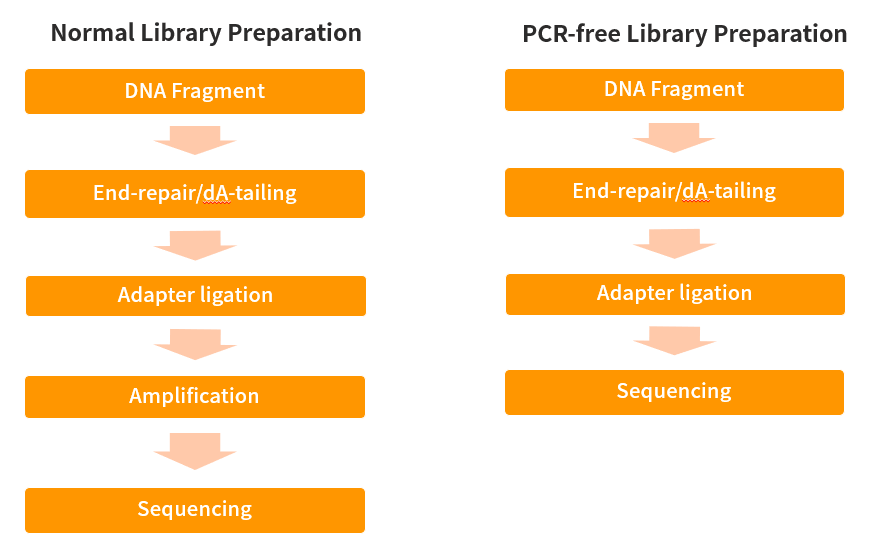
Figure 1. Flow chart of conventional library construction and PCR-Free library construction
2. What are the advantages of PCR-Free technology?
In the routine library construction process, the amplification enzyme may introduce errors. Through cyclic amplification, these errors will be accumulated and amplified, reducing the fidelity of DNA sequence replication. In addition, DNA polymerase also has a certain amplification bias, especially for regions with large differences in GC content or secondary structures, resulting in low amplification efficiency and poor coverage uniformity; while introducing wrong InDels, it will also bring a large number of Duplication causes waste of DNA data volume and increases sequencing costs. The PCR-Free technology can well avoid the above-mentioned problems introduced by PCR amplification in conventional library construction. In the process of library construction and sequencing, if there is a PCR amplification process, it will have a greater impact on the uniformity of genome coverage. For DNA templates, some have complex secondary structures and some have large differences in thermal stability. These factors will affect the efficiency of PCR amplification. Therefore, in the PCR amplification process, it cannot be guaranteed that all genomic fragments can obtain the same amplification efficiency, but there is an obvious amplification bias, such as low coverage in high GC or low GC regions of the genome. However, there is no PCR in the whole process of PCR-Free sequencing. Compared with PCR library construction, the coverage of high GC regions and AT/TA repeat regions of the genome is improved. The specific advantages are as follows.
2.1 Streamlined library preparation process, saving time
PCR-Free library preparation eliminates the need for PCR amplification and other steps, streamlining the library preparation process and saving time.
2.2 Reduced library preparation cost
In the conventional library preparation process, expensive high-fidelity enzymes are used for PCR amplification to ensure sequence authenticity. At the same time, PCR-Free eliminates the PCR amplification step, reducing library preparation costs.
2.3 Reduce amplification bias
DNA amplification enzymes have certain amplification biases, especially for templates with significant differences in GC content or secondary structure. Therefore, PCR-Free can effectively avoid the amplification bias caused by the polymerase.
2.4 No PCR Duplication
PCR-Free can reduce Duplicate reads and increase Unique Mapping rates and effective data utilization.
2.5 Reduced Error Pare
PCR amplification is more likely to introduce replication errors of Insertion & Deletion, resulting in a higher Error Rate of Indel sequencing, while PCR-Free can effectively avoid the generation and accumulation of such errors.
3. Data display of PCR-Free
Hieff NGS™ Ultima Pro PCR Free DNA Library Prep Kit V2 (Cat# 12196ES) has a better library conversion rate than competing products.
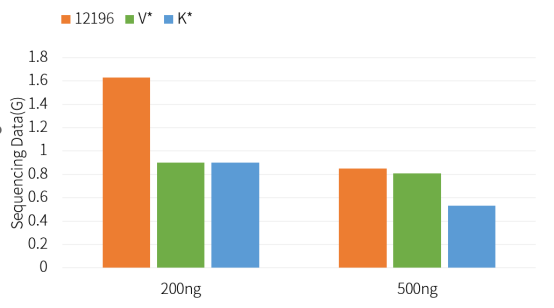
Figure 2. Quantity of off-machine sequencing data for PCR-Free library construction
4. FAQ
Q: What types of samples are suitable for PCR-free libraries?
A: Some samples with complex secondary structures, high GC, and PCR preference can choose a PCR-free library to construct.
Q: What is the general fragment size of the PCR-free library?
A: The PCR-free library itself has no special requirements on the fragment size. The PCR product library, library is constructed according to the size of the PCR product. For the genomic library, it is recommended to be around 350bp, so that the quality of the library data is relatively good.
5. Product information
The product that Yeasen can provide is shown in Table 1.
Table 1. Product information
| Product Name | Cat. No. | Size |
| Hieff NGS™ Ultima Pro PCR Free DNA Library Prep Kit V2 | 12196ES24/96 | 24/96T |
6. Regarding reading
The solution for the NGS library preparation from DNA samples
About NGS-related technology, how much do you know?
