Description
Hifair™ Ⅲ 1st Strand cDNA Synthesis SuperMix for qPCR (gDNA Digester Plus) is a readjust premix developed based on Hifair™ⅢReverse Transcriptase. Compared with Hifair™ ⅡReverse Transcriptase, Hifair™ Ⅲ Reverse Transcriptase has significantly higher thermal stability and can withstand reaction temperatures up to 60℃, and is suitable for Reverse transcription of RNA templates with complex secondary structures. At the same time, the enzyme enhances the affinity with templates, which is ideal for the reverse transcription of a small number of templates and low-copy genes.
The premix contains 5×gDNA Digester Mix and 4×Hifair™ Ⅲ SuperMix Plus. 5×gDNA Digester Mix can remove residual genomic DNA contamination in RNA templates to ensure more reliable follow-up results. 4×Hifair™ Ⅲ SuperMix Plus contains all components required for Reverse Transcriptase (Buffer, dNTP, Hifair™ Ⅲ Reverse Transcriptase, RNase inhibitor, Random Primers/Oligo (dT)18 Primer Mix), RNA template and Rnase-free H2O are added to reverse transcription, and gDNA digester is terminated simultaneously to ensure the integrity of cDNA.
The product is suitable for two-step RT-QPCR detection, specially optimized for qPCR, proportionally optimized Random Primers/Oligo (dT)18 Primer Mix, enabling cDNA synthesis to be initiated from all regions of RNA transcript with the same reverse transcription efficiency. The authenticity and repeatability of qPCR results are guaranteed to the greatest extent. The reverse transcription product is compatible with SYBR Green method and TaqMan probe method qPCR, Hifair™ UNICON™ qPCR SYBR Green Master Mix, Hifair™ qPCR SYBR Green Master Mix or Hifair™ qPCR TaqMan Probe Master Mix can be selected according to experimental purposes, is used in combination to perform high-performance gene expression analysis.
Features
- High-performance reverse transcription
- Increase reaction temperature - First strand cDNA can be synthesized in the temperature range of 55 to 60°C
- Efficient removal of genomic residues
- Broad linear detection range
Applications
- Construction of cDNA library
- Antisense RNA synthesis
Specifications
| Type of end product | cDNA (First-Strand) |
| PCR method | RT-PCR |
| Type of reagent | Reverse Transcription |
| Type of sample | RNA |
| Optimal reaction temperature | 55℃-60℃ |
| Reverse transcriptase | M-MLV |
Components
| Components No. | Name | 11141ES10 (10T) | 11141ES60 (100T) |
| 11141-A | RNase-free H2O | 1 mL | 2×1 mL |
| 11141-B | 5×gDNA digester Mix | 30 μL | 300 μL |
| 11141-C | 4×HifairTM Ⅲ SuperMix plus | 50 μL | 500 μL |
Shipping and Storage
The product is shipped with dry ice and can be stored at -15℃ ~ -25℃ for 18 months.
Figures
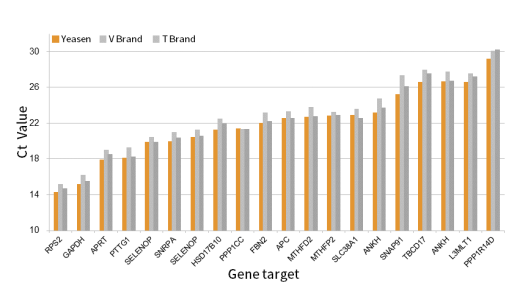
Figure 1. Excellent reverse transcription efficiency, suitable for genes with different expression abundances with different GC contents
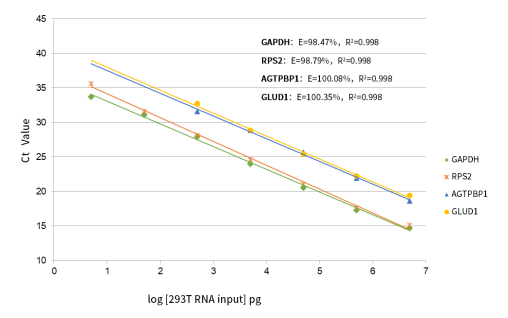
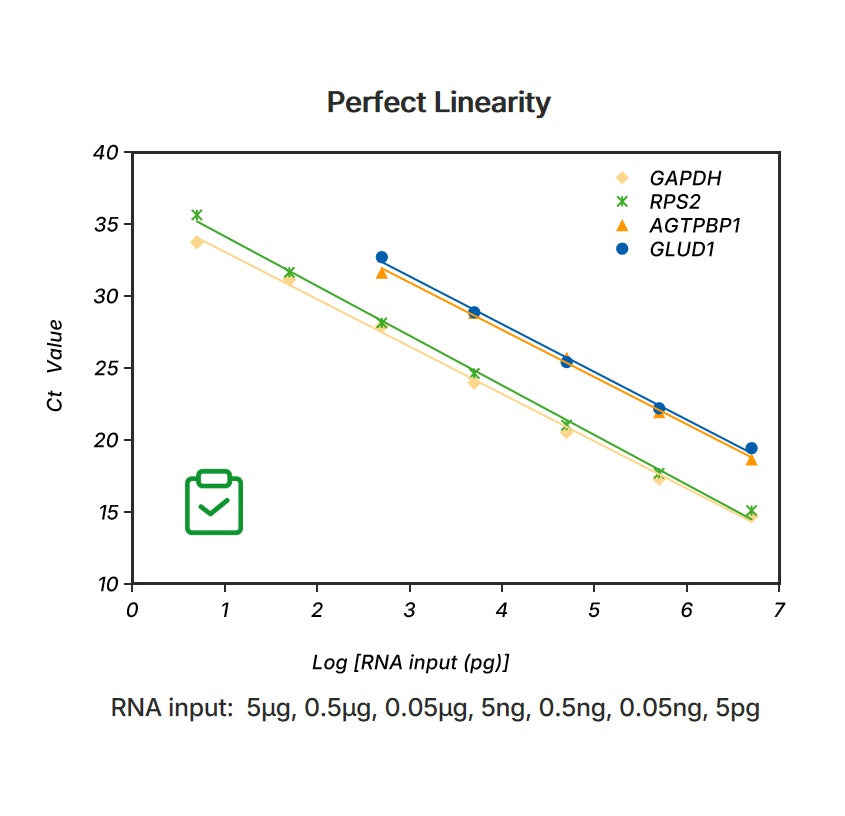
Figure 2. Linear assay range Total RNA 5 pg~5 μg
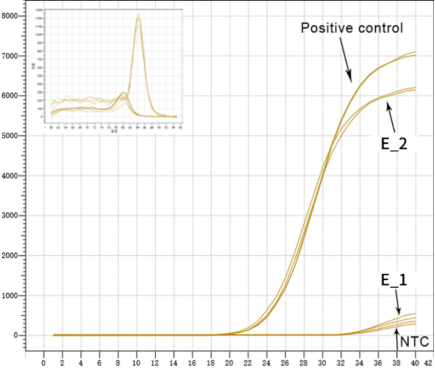
Figure 3. Efficient removal of genomic residues
Cited from"An RNA-cleaving threose nucleic acid enzyme capable of single point mutation discrimination. Nat Chem . 2022 Mar;14(3):350-359. doi: 10.1038/s41557-021-00847-3."
Cited from"The tetratricopeptide repeat protein OsTPR075 promotes heading by regulating florigen transport in rice. Plant Cell . 2022 Sep 27;34(10):3632-3646. doi: 10.1093/plcell/koac190."
[1] Yuan B, Peng Q, Cheng J, et al. Structure of the Ebola virus polymerase complex. Nature. 2022;610(7931):394-401. doi:10.1038/s41586-022-05271-2(IF:69.504)
[2] Bi Q, Wang C, Cheng G, et al. Microglia-derived PDGFB promotes neuronal potassium currents to suppress basal sympathetic tonicity and limit hypertension. Immunity. 2022;55(8):1466-1482.e9. doi:10.1016/j.immuni.2022.06.018(IF:43.474)
[3] You XJ, Zhang S, Chen JJ, et al. Formation and removal of 1,N6-dimethyladenosine in mammalian transfer RNA. Nucleic Acids Res. 2022;50(17):9858-9872. doi:10.1093/nar/gkac770(IF:19.160)
[4] Ni X, Wu W, Sun X, et al. Interrogating glioma-M2 macrophage interactions identifies Gal-9/Tim-3 as a viable target against PTEN-null glioblastoma. Sci Adv. 2022;8(27):eabl5165. doi:10.1126/sciadv.abl5165(IF:14.957)
[5] Chen Y, Shen J, Zhang L, et al. Nuclear translocation of OsMFT1 that is impeded by OsFTIP1 promotes drought tolerance in rice. Mol Plant. 2021;14(8):1297-1311. doi:10.1016/j.molp.2021.05.001(IF:13.164)
[6] Yang L, Chen Y, Xu L, et al. The OsFTIP6-OsHB22-OsMYBR57 module regulates drought response in rice. Mol Plant. 2022;15(7):1227-1242. doi:10.1016/j.molp.2022.06.003(IF:13.164)
[7] Song JM, Xie WZ, Wang S, et al. Two gap-free reference genomes and a global view of the centromere architecture in rice. Mol Plant. 2021;14(10):1757-1767. doi:10.1016/j.molp.2021.06.018(IF:13.164)
[8] Lin QY, Zhang YL, Bai J, Liu JQ, Li HH. VEGF-C/VEGFR-3 axis protects against pressure-overload induced cardiac dysfunction through regulation of lymphangiogenesis. Clin Transl Med. 2021;11(3):e374. doi:10.1002/ctm2.374(IF:11.492)
[9] Zhang L, Zhang F, Zhou X, et al. The tetratricopeptide repeat protein OsTPR075 promotes heading by regulating florigen transport in rice [published online ahead of print, 2022 Jun 28]. Plant Cell. 2022;koac190. doi:10.1093/plcell/koac190(IF:11.277)
[10] Mao Z, Xiao H, Shen P, et al. KRAS(G12D) can be targeted by potent inhibitors via formation of salt bridge. Cell Discov. 2022;8(1):5. Published 2022 Jan 25. doi:10.1038/s41421-021-00368-w(IF:10.849)
[11] Li M, Chen J, Liu Y, et al. Rational design of AAVrh10-vectored ACE2 functional domain to broadly block the cell entry of SARS-CoV-2 variants. Antiviral Res. 2022;205:105383. doi:10.1016/j.antiviral.2022.105383(IF:10.103)
[12] Luo F, Yang W, Yin M, et al. A chromosome-level genome of the human blood fluke Schistosoma japonicum identifies the genomic basis of host-switching. Cell Rep. 2022;39(1):110638. doi:10.1016/j.celrep.2022.110638(IF:9.423)
[13] Wang X, Lin C, Wu S, et al. Cannabidivarin alleviates neuroinflammation by targeting TLR4 co-receptor MD2 and improves morphine-mediated analgesia. Front Immunol. 2022;13:929222. Published 2022 Aug 10. doi:10.3389/fimmu.2022.929222(IF:8.786)
[14] Jiang R, Su G, Chen X, et al. Esculetin inhibits endometrial cancer proliferation and promotes apoptosis via hnRNPA1 to downregulate BCLXL and XIAP [published online ahead of print, 2021 Sep 1]. Cancer Lett. 2021;521:308-321. doi:10.1016/j.canlet.2021.08.039(IF:8.679)
Payment & Security
Your payment information is processed securely. We do not store credit card details nor have access to your credit card information.
Inquiry
You may also like
FAQ
The product is for research purposes only and is not intended for therapeutic or diagnostic use in humans or animals. Products and content are protected by patents, trademarks, and copyrights owned by Yeasen Biotechnology. Trademark symbols indicate the country of origin, not necessarily registration in all regions.
Certain applications may require additional third-party intellectual property rights.
Yeasen is dedicated to ethical science, believing our research should address critical questions while ensuring safety and ethical standards.