Work along both lines, miRNA expression analysis made easier
The length of mature miRNA is only about 20nt, usually, the forward primer will cover its full length or even have an excess part, and the reverse primer will have nowhere to be placed. The current market solution is to increase the length of the reverse transcript during reverse transcription. Common methods include the tailing method and the stem-loop method.
1. Introduction
2. Products Recommended
3. Related product list
1. Introduction
MicroRNAs (miRNAs) are single-stranded small RNAs with a size of about 21-23 bases, which are generated from single-stranded RNA precursors with a hairpin structure of about 70-90 bases after Dicer enzyme processing. miRNA mainly binds specifically to the 3'-end non-coding region of target gene mRNA, and regulates the translation process of post-transcriptional target gene mRNA, thereby blocking mRNA translation and promoting mRNA degradation, resulting in decreased protein expression. The role of miRNAs is involved in the occurrence and development of various cardiovascular diseases such as ontogeny, tissue differentiation, cell proliferation, and apoptosis.
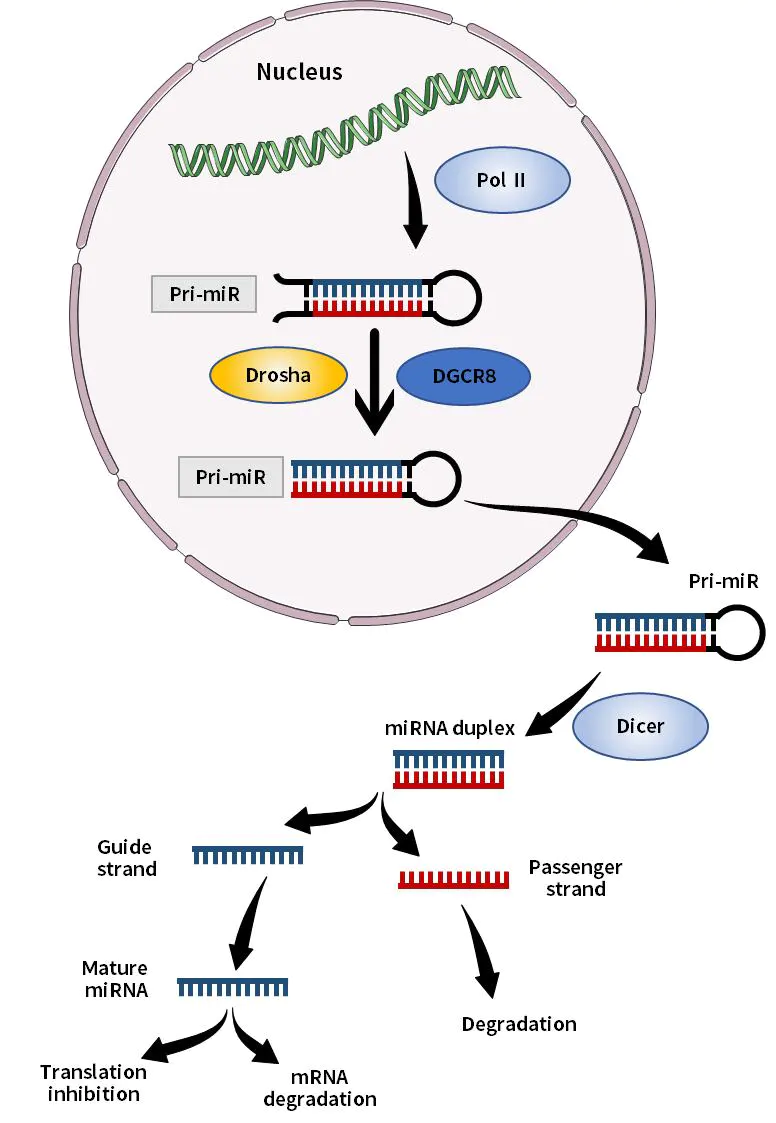
Figure 1. Schematic diagram of miRNA synthesis in vivo
The length of mature miRNA is only about 20nt, usually, the forward primer will cover its full length or even have an excess part, and the reverse primer will have nowhere to be placed. The current market solution is to increase the length of the reverse transcript during reverse transcription. Common methods include the tailing method and the stem-loop method.
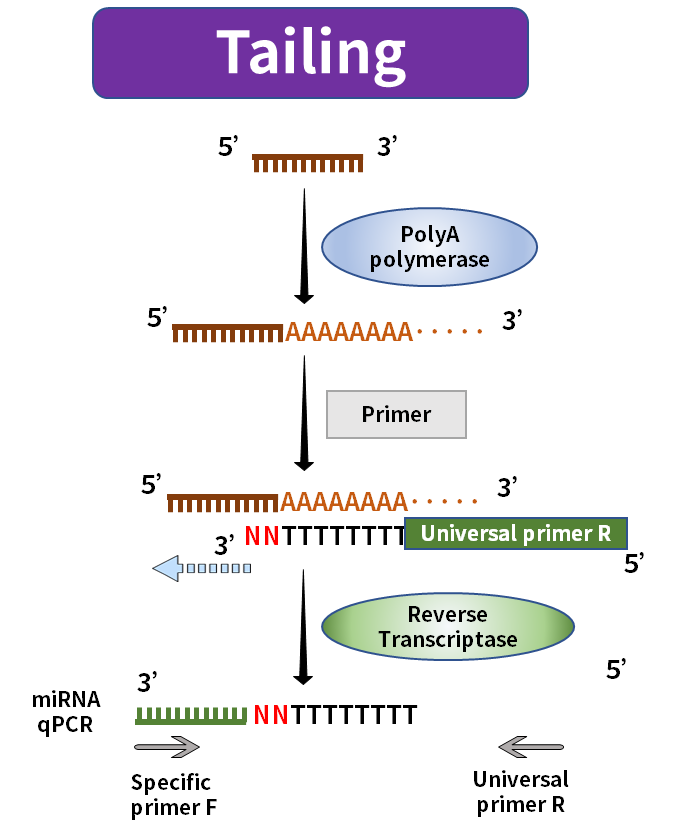
Figure 2. miRNA synthesis by tailing method
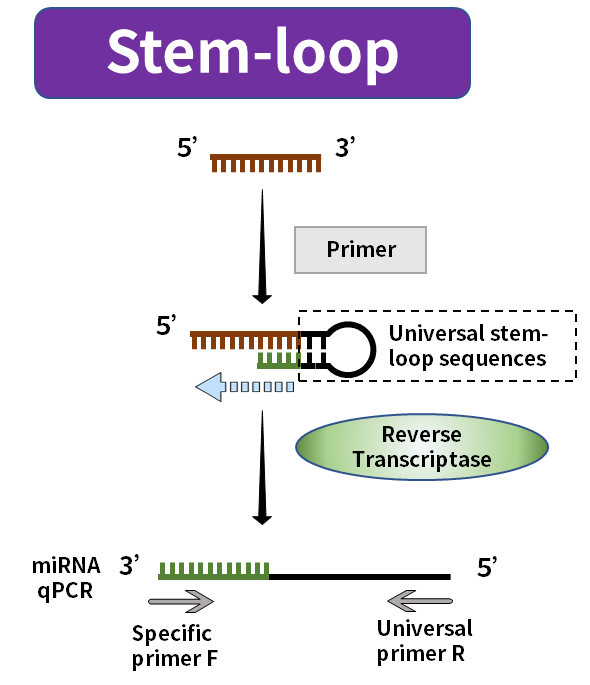
Figure 3. miRNA synthesis by the stem-loop method
The tailing method uses PolyA polymerase to add Poly(A) tail to mature miRNA and uses Oligo-dT with the universal sequence as a reverse transcription primer to synthesize the lengthened miRNA corresponding to the first strand of cDNA. The stem-loop method uses the universal sequence folded into the stem-loop structure as a primer to synthesize the first strand of the cDNA corresponding to the elongated miRNA. The stem-loop structure will be opened at a suitable temperature during the subsequent amplification process and combined with the relevant amplification primers.
Table 1. Comparison of Tailing and Stem-loop
|
Category |
Tailing |
Stem-loop |
|
Applicability |
Suitable for studying multiple different miRNAs simultaneously |
Suitable for accurate quantitative detection of a few miRNAs in a sample |
|
Advantage |
A single reverse transcription can detect multiple miRNAs with high throughput |
Only one miRNA can be detected by reverse transcription at a time, with strong specificity |
|
System |
Together with the miRNA involved, reverse transcription in the same reaction system with high accuracy |
Internally involved miRNAs are separated and reverse transcribed in the two systems, which is prone to errors |
|
Specificity |
★★★★☆ |
★★★★★ |
|
Sensitivity |
★★★★★ |
★★★★☆ |
|
Time |
★★★★☆ |
★★★★★ |
2. Products Recommended
2.1 Yeasen miRNA tailed inversion quantitative combination (11148ES&11171ES)
Figure 4. 11148ES-Hifair™ miRNA 1st Strand cDNA Synthesis Kit
The kit adopts the tailing method to reverse transcription of miRNA first-strand cDNA. The 2×Hifair™ miRNA RT buffer in the product contains all the raw materials and primers for miRNA tailing reaction and reverse transcription reaction, and has been carefully optimized. It can ensure that the poly(A) modification and reverse transcription processes of the miRNA 3' end are carried out efficiently simultaneously.
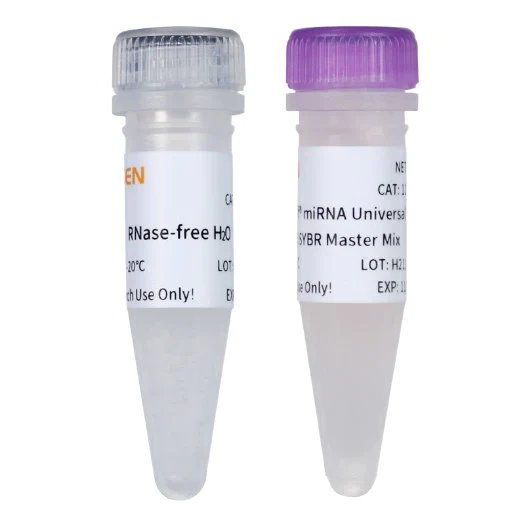
Figure 5. 11171ES -Hieff™ miRNA Universal qPCR SYBR Master Mix
A new generation of premixed fluorescence quantitative PCR detection reagents specially developed for miRNA quantitative detection, containing special ROX Passive Reference Dye, suitable for all qPCR instruments, no need to adjust the concentration of ROX on different instruments, just in the preparation reaction Amplification can be performed by adding primers and templates to the system. The DNA polymerase adopts chemically modified hot-start polymerase, and cooperates with a special Buffer system, which makes the reaction specificity and sensitivity higher, and can accurately quantify in a wider range.
2.2 Product performance
2.2.1 Compatible with all-platform qPCR instruments

Figure 6. Compatible results with different qPCR instruments
Using the Total RNA of 293T cells as a template, reverse transcription was performed with Hifair™ miRNA 1st Strand cDNA Synthesis Kit (Cat# 11148ES). The obtained cDNA was diluted 10-fold, and then used Hieff™ miRNA Universal qPCR SYBR Master Mix (Cat# 11171ES) to detect the internal reference genes of hsa-miR-let-7b-5p, hsa-miR-let-7c-5p, and U6, respectively.
2.2.2 High detection rate, up to 10 pg
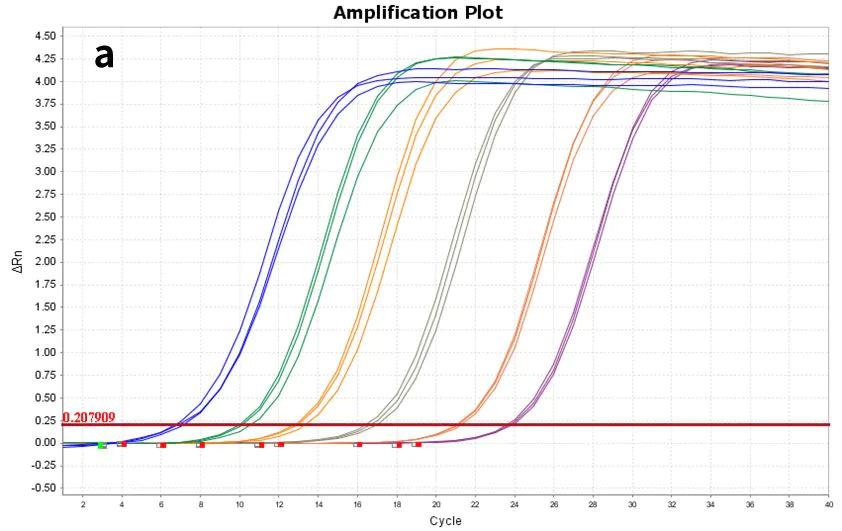
Figure 7. Amplification plot
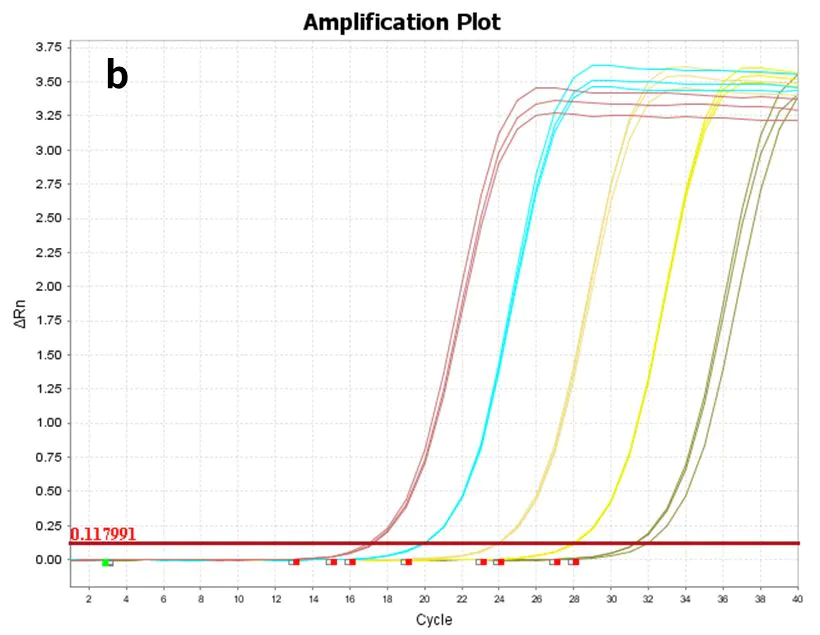
Figure 8. Amplification plot
Using synthetic hsa-miR-let-7e-5p(a) and 293T cell Total RNA(b) as templates, diluted to the following gradients, respectively: 60 copies to 606 copies (6 gradients) and 10 pg-100 ng (5 gradients), reverse transcribed with Hifair™ miRNA 1st Strand cDNA Synthesis Kit (Cat# 11148ES), and the resulting cDNA was detected with Hieff™ miRNA Universal qPCR SYBR Master Mix (Cat# 11171ES) for hsa-miR-let-7e -5p gene expression.
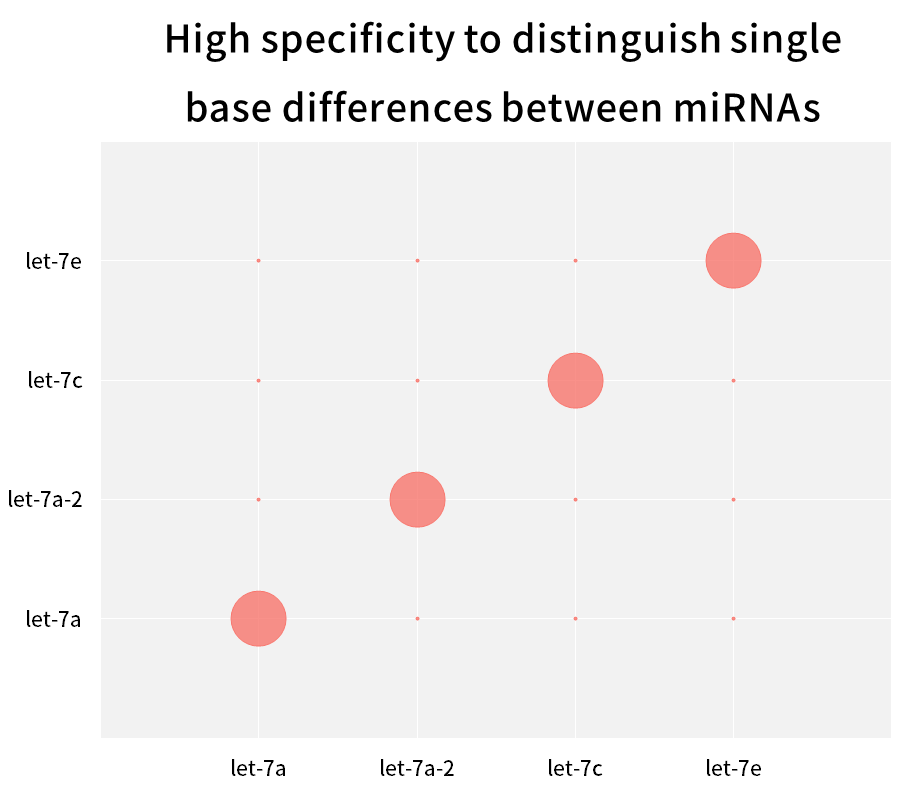
2.2.3 High specificity, able to distinguish single base differences between miRNAs in the same family
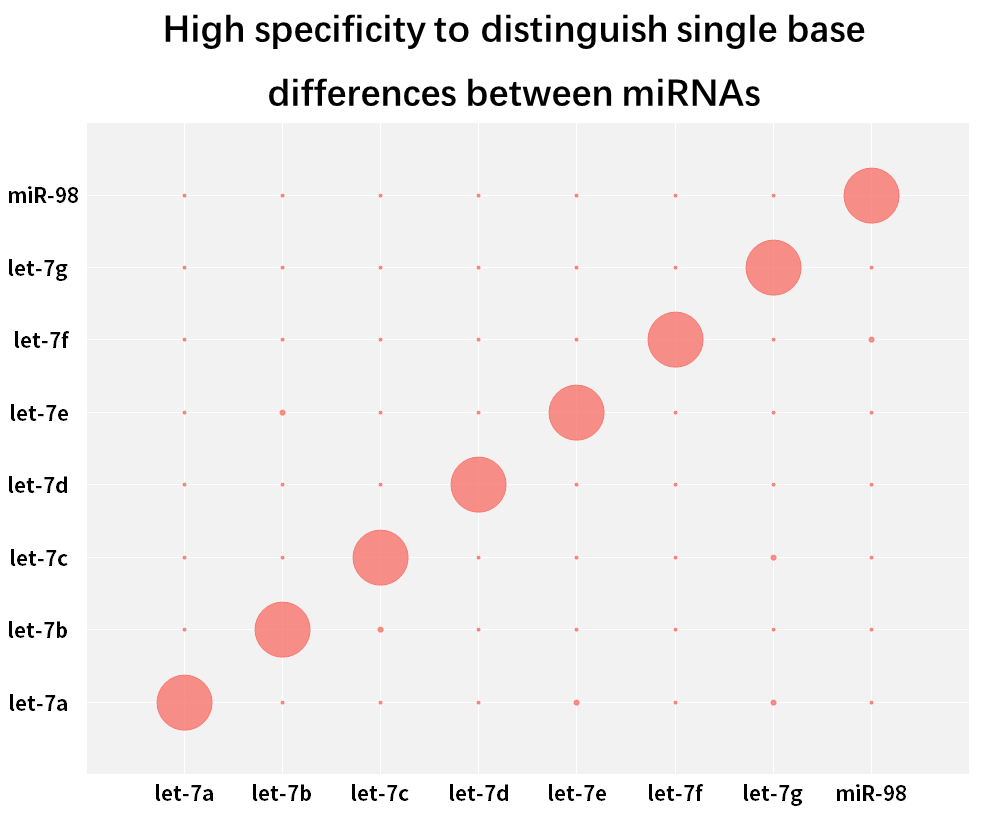
Figure 9. Distinguish single base differences between miRNAs
The human hsa-let-7 family has multiple miRNAs, with few bases that differ from each other, or even only a single base difference. Synthetic miRNAs for each Let-7 isoform (hsa-miR-let-7a~Let-7i, has-miR-98) were reverse transcribed into cDNA using the Hifair™ miRNA 1st Strand cDNA Synthesis Kit (Cat# 11148ES) , using Hieff™ miRNA Universal qPCR SYBR Master Mix (Cat# 11171ES) to amplify these miRNAs separately with different primers, and calculate the match rate using Equation 2-ΔCT x 100%. The results show that closely related subtype family members can be effectively distinguished.
2.2.4 Excellent amplification efficiency
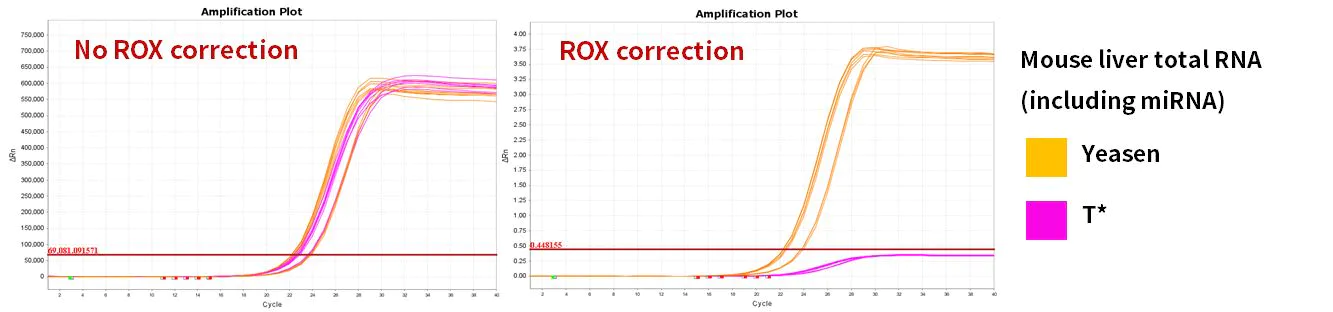
Figure 10. Amplification efficiency
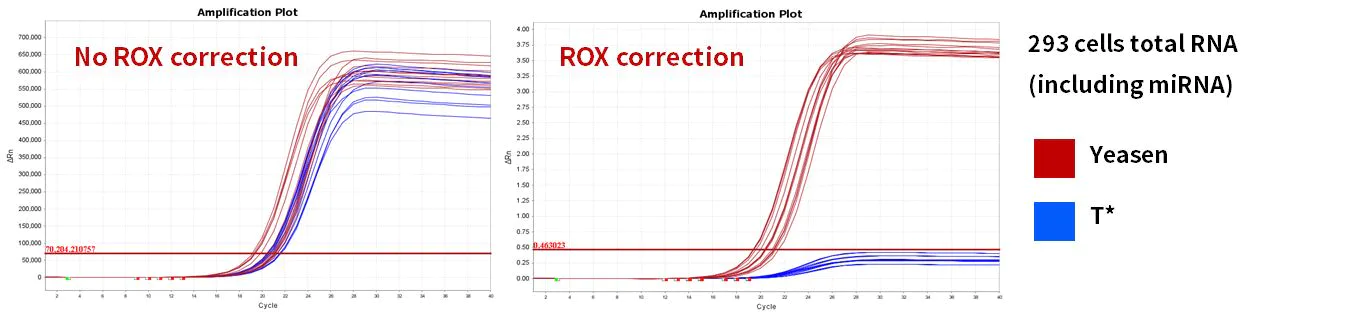
Figure 11. Amplification efficiency
miR-let-7b-5p, miR-let-7c-5p, miR-let-7e-5p, miR-let-7f-5p genes were amplified using human 293T cells and mouse liver Total RNA as templates. The results showed that compared with similar products, Hifair™ miRNA 1st Strand cDNA Synthesis Kit (Cat# 11148ES) matched the reaction system of Hieff™ miRNA Universal qPCR SYBR Master Mix (Cat# 11171ES) with excellent performance.
2.2.5 Excellent stability

Figure 12. Product stability analysis at 37℃ for 7 days

Figure 13. Analysis of results after 50 freeze-thaw cycles
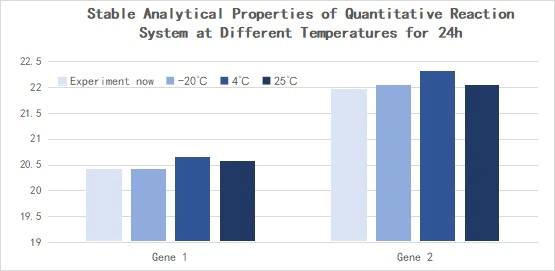
Figure 14. Stable analytical properties of the quantitative reaction system at different temperatures for 24h
Reverse transcription was performed with Hifair™ miRNA 1st Strand cDNA Synthesis Kit (Cat# 11148ES) using 293T cell Total RNA as template, and the obtained cDNA was diluted 10-fold and detected with Hieff™ miRNA Universal qPCR SYBR Master Mix (Cat# 11171ES). Expression of hsa-miR-let-7b-5p, hsa-miR-let-7c-5p, hsa-miR-let-7e-5p. △Ct<0.5.
2.3 Yeasen miRNA stem-loop inversion quantitative combination (11139ES&11170ES)
Figure 15. 11139ES-Hifair™ III 1st Strand cDNA Synthesis Kit(gDNA digester plus)
The kit was developed based on Hifair™ III Reverse Transcriptase. The enzyme cDNA synthesis speed is fast, and the thermal stability is greatly improved. At the same time, the enzyme enhances the affinity with the template and is suitable for the reverse transcription of a small amount of templates and low-copy genes. The ability of Hifair™ III Reverse Transcriptase to synthesize full-length cDNA has also been improved, allowing the synthesis of cDNA up to 19.8 kb.

Figure 16. 11170ES-Hieff™ miRNA Universal qPCR SYBR Master Mix
This product is a special master mix for miRNA quantification using SYBR Green I chimeric fluorescence method. Because miRNA sequences are short and miRNA sequences in the same family are often highly similar, specificity is extremely required during quantification. This product is based on hot-start DNA polymerase, with optimized Buffer, which can greatly reduce non-specific amplification; at the same time, the special ROX Reference Dye makes the master mix suitable for all qPCR instruments, and there is no need to adjust ROX on different instruments Amplification can be performed by adding primers and templates when preparing the reaction system.
2.3.1 Compatible with all-platform qPCR instruments
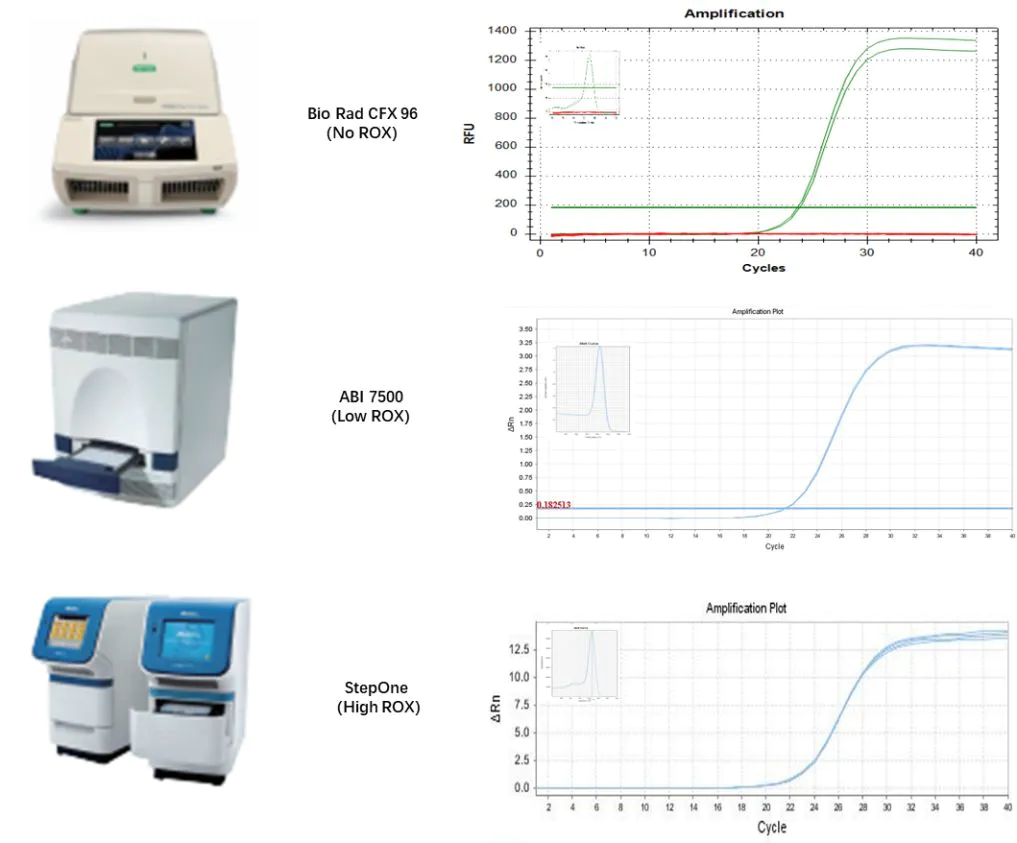
Figure 17. Compatible results with different qPCR instruments
Using total RNA of mouse liver cells as a template, reverse transcription was performed with Hifair™ III 1st Strand
cDNA Synthesis Kit (gDNA digester plus) (Cat# 11139ES), and the obtained cDNA was diluted 40-fold and used Hieff™ miRNA Universal qPCR SYBR Master Mix (Cat# 11170ES) was used to amplify the human mmu-miR-125a gene, respectively.
2.3.2 High sensitivity, up to 10 pg

Figure 18. Sensitivity detection
Using total RNA of mouse liver cells as a template, reverse transcription was performed with Hifair™ III 1st Strand cDNA Synthesis Kit (gDNA digester plus) (Cat# 11139ES), and the obtained cDNA was serially diluted 10-fold (range 10pg/μL). -0.1 μg/μL), amplify the mmu-miR-125a, mmu-miR-132, and mmu-miR-351 genes of murine origin.
2.3.3 High specificity, able to distinguish single base differences between miRNAs in the same family
Figure 19. Distinguish single base differences between miRNAs
The human hsa-let-7 family has multiple miRNAs, with few bases that differ from each other, or even only a single base difference. These miRNAs were amplified separately with different primers using Hieff™ miRNA Universal qPCR SYBR Master Mix (Cat# 11170ES), and the matching rate was calculated using the formula 2^-Δct x 100%.
2.3.4 Good repeatability

Figure 20. Repeatability detection
Using the Total RNA of human 293T cells as a template for reverse transcription, reverse transcription was performed with Hifair™ III 1st Strand cDNA Synthesis Kit (gDNA digester plus) (Cat# 11139ES), and the obtained cDNA was diluted 50-fold and used Hieff™ miRNA Universal qPCR SYBR Master Mix (Cat# 11170ES) was used to amplify the human hsa-let-7a gene in a 90-well replicate experiment.
2.3.5 Good stability

Figure 21. Stability analysis of 11170ES at different temperatures for 14 days
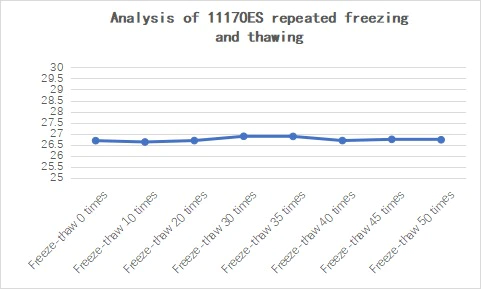
Figure 22. Analysis of 11170ES repeated freezing and thawing

Figure 23. Stable analysis properties of the quantitative reaction system at different temperatures for 24h
The total RNA of mouse liver cells was used as a template for reverse transcription with Hifair™ III 1st Strand cDNA Synthesis Kit (gDNA digester plus) (Cat# 11139ES). The obtained cDNA was diluted 100-fold to amplify the murine mmu- miR-132 gene.
3. Related product list
Yeasen Biotechnology (Shanghai) Co., Ltd., founded in 2014, is a high-tech enterprise engaged in the R &D and production of tool enzyme raw materials and antigen antibodies. Its products include molecular diagnostic enzymes, proteins, and antibodies used in pharmaceuticals, food safety testing, breeding, justice, and other industries. We are committed to providing customers in the field of life sciences with high-quality products and services. The related products provided by Yeasen are as follows:
Table 2. Related product list