Various types of magnetic beads in NGS: DNA\RNA\mRNA magnetic beads
Magnetic beads are one of the necessary products in the construction of high-throughput sequencing libraries. They can purify DNA or RNA, screen target-size DNA fragments, and enrich target nucleic acids. With the development of sequencing technology, magnetic beads specialized in different application fields have emerged, including DNA purification magnetic beads, DNA size selection magnetic beads, RNA Purification magnetic beads, and mRNA enrichment magnetic beads. So what are the principles of various magnetic beads? How to choose?
Request Free Sample and Lower Price for Bulk Purchasing
1. DNA beads
2. Star DNA beads introduction
3. RNA purification beads
4. mRNA-enriched magnetic beads
5. Yeasen magnetic beads product recommendation
1. DNA Beads
1.1 Basic principles
The principle of purification of nucleic acid by magnetic beads is based on solid-phase reversible immobilization (SPRI). Under certain conditions, the nucleic acid is selectively bound to the magnetic beads, while pollutants remain in the solution. After applying a magnetic field, the magnetic beads binding with the target molecules are separated from the solution, and then the magnetic beads are cleaned to further remove pollutants. Because the combination of magnetic beads and nucleic acid molecules is reversible, the nucleic acid can be eluted from the magnetic beads with a low salt buffer.
The outer surface of magnetic beads is modified with silicon hydroxyl or carboxyl functional groups. In the purification buffer system containing PEG and high salt ions, DNA is bound with beads by forming an ion bridge of DNA salt ion carboxyl group. This binding is reversible. The ion bridge is removed in the TE buffer without PEG and salt ions, and finally, DNA is purified. Based on carboxyl magnetic beads, different magnetic bead inputs and purification buffers will bind different fragment sizes. The magnetic beads preferentially bind large fragments of DNA, so in the first round, magnetic beads are used to bind the fragments larger than the target fragment, and the supernatant is retained; In the second round, the magnetic beads bind the target fragment in the supernatant and then discard the supernatant; Finally, the target fragment is eluted from the magnetic beads, which the DNA fragments are the target size.
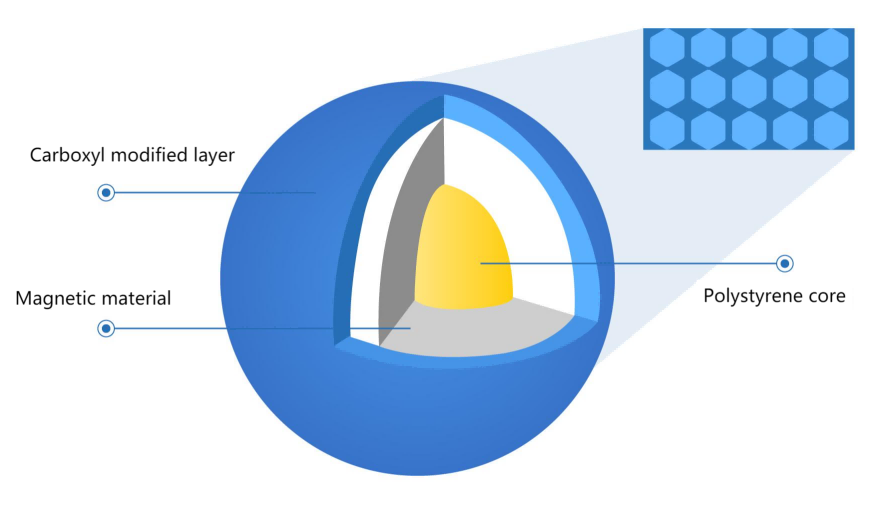
Figure 1. Carboxyl magnetic bead structure
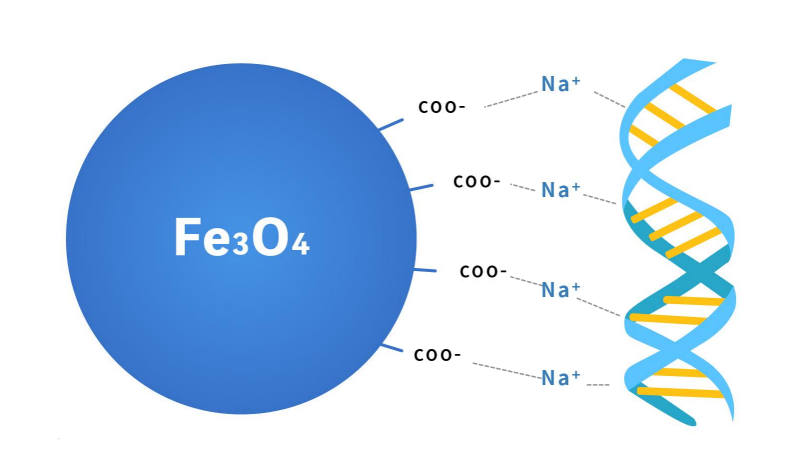
Figure 2. Principle of DNA purification magnetic beads
1.2 Operation Process
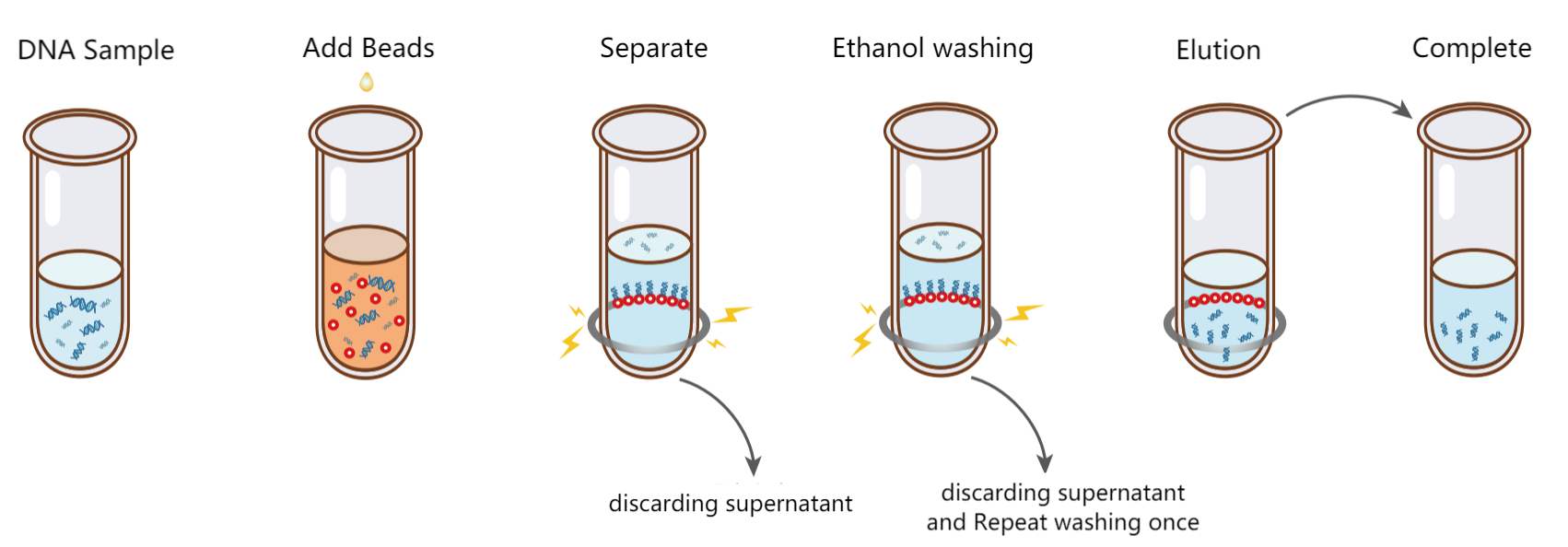
Figure 3. DNA purification steps
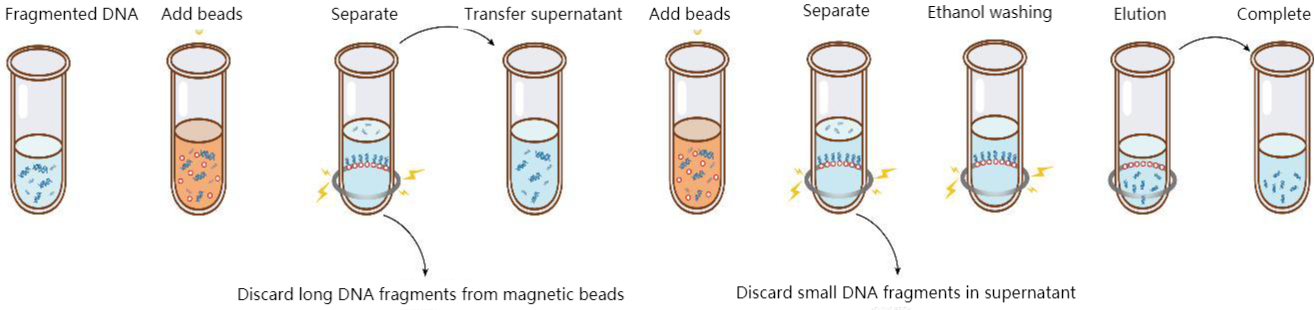
Figure 4. DNA size selection steps
1.3 Tips for using magnetic beads
Improved yield and accurate size selection
(1) Mix well before use, and equilibrate the magnetic beads to room temperature for at least 30min.
——The solubility of PEG in the magnetic beads buffer is easily affected by pH and temperature. Before use, it is necessary to equilibrate to room temperature to make the magnetic beads evenly suspended and PEG fully dissolved to avoid affecting the adsorption and separation effects
(2) The adsorption time should be sufficient, and fully mixed to make the adsorption adequate;
(3) Washing with 80% ethanol during purification or separation;
——Under 80% ethanol, the nucleic acid is dehydrated and gathered closely, and will not dissolve. The residual enzymes, buffers, impurities, etc. in the system are cleaned with ethanol to obtain a purer library.
(4) During size selection, confirm the sample volume to ensure that the magnetic bead addition ratio is correct;
——During size selection, it is necessary to accurately input the corresponding volume of magnetic beads, so that the proportion is accurate, because the change of PEG and salt ion concentration in the buffer may affect the results.
(5) Before the elution step, let the alcohol volatilize completely, but prevent the beads from over-drying
——Generally, it can be dried in 2~3min. When the number of magnetic beads is large and difficult to dry, It is recommended to operate in multiple centrifuge tubes.
(6) If the magnetic beads are agglomerated or slabbed, the elution time can be extended after fully mixing;
——It may be caused by impurities in DNA or too long drying time. Generally, when there are many impurities in DNA, it is recommended to purify it first before size selection.
2. Star DNA beads introduction
Hieff NGS™ DNA selection beads are based on the principle of SPRI (solid phase reverse immobilization), using imported magnetic beads raw materials, and an optimized buffer system, which can be used for DNA fragment size selection and purification during the construction of the NGS library. It is used in the same way as the used AMPure XP beads, and the fragment recovery efficiency and library size distribution are highly consistent with them.
2.1 Introduction of purification performance
- Unique buffer system: DNA fragments down to 50bp can be recovered.
- High recovery rate: ≥90%.
- Effectively remove impurities: Effectively remove impurities such as primer dimer, dNTP, inorganic salt, and protein.
- Wide application range: Suitable for DNA purification in such scenarios as enzyme digestion, ligation, cloning, and NGS library building.
Table 1. DNA purification and recovery efficiency of magnetic beads with different ratios
| Experience Group | Recovery concentration of magnetic beads in this batch(ng/μL) | Recovery Rate | AMPure XP beads Group(ng/μl) ④ | Recovery Rate | CV% | |||
| Ratio | ① | ② | ③ | Average | ||||
| 1.8× | 22.2 | 23.4 | 22.8 | 22.8 | 96.92% | 22.2 | 94.37% | 2.55% |
| 0.8× | 20.2 | 19.3 | 18.5 | 19.33 | 82.19% | 17.8 | 75.67% | 6.52% |
| 0.6× | 16.3 | 17.3 | 16.8 | 16.8 | 71.42% | 16.2 | 68.87% | 2.55% |
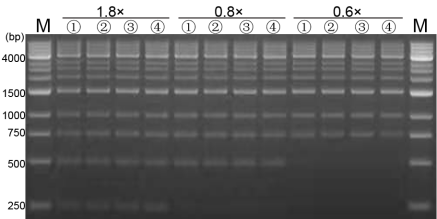
Figure 5. Purification effect of magnetic beads agarose gel electrophoresis results
M:1kb DNA ladder;
2.2 Size selection performance
- Easy to operate: the separation principle and experimental operation are completely consistent with XP magnetic beads
- Accurate size selection: the size of sorted fragments is accurate and stable
- Wide applicability: suitable for building DNA and RNA libraries and adapting to fragment size selection of various sample types
- Ultra-high cost performance: stable quality, more economical price, more considerate pre-sales and after-sales service
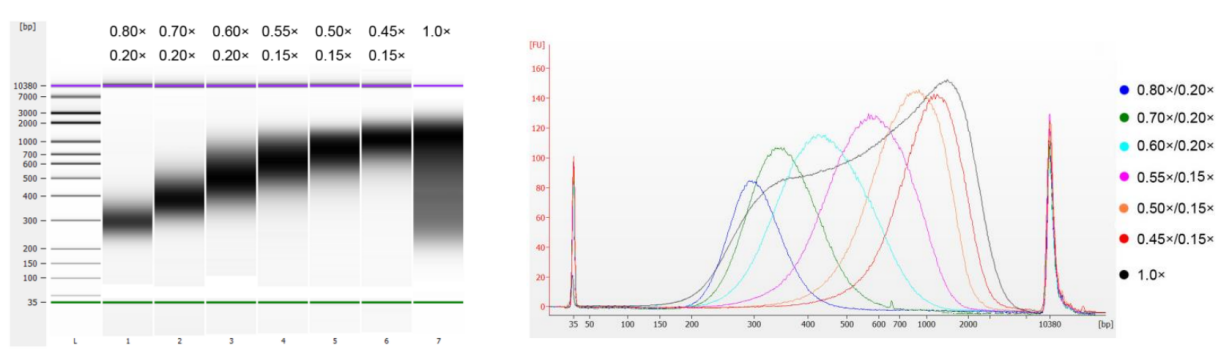
Figure 6. DNA Size selection results
3. RNA Purification beads
Hieff NGS™ RNA cleaner is based on the SPRI principle, which can efficiently remove proteins, salt ions, and other impurities to obtain high-purity concentrated RNA samples, and carefully optimize the acidic buffer system to maintain the stability of RNA structure and prevent RNA degradation. It is suitable for RNA library construction, purification of total RNA samples after rRNA removal, purification of RNA products transcribed in vitro, purification of RNA labeled products, purification of synthetic RNA, etc.
- High quality: imported raw materials, highly stable quality
- Optimized buffer system: ensure the purity and integrity of RNA and effectively prevent RNA degradation
- High efficiency: efficient recovery, suitable for RNA library construction or in vitro transcription product recovery, etc.
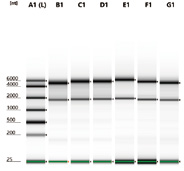
Figure 7. Electrophoretogram of different samples after purification
4. mRNA-enriched magnetic beads
4.1 Principle of mRNA isolation
mRNA separation and enrichment magnetic beads are magnetic beads that surface modified with oligo (dT). Through the principle of hybridization, they can specifically bind mRNA with Poly A tail and specifically separate mRNA from total RNA or tissue cells. This kit with the optimized product formula can efficiently isolate high-purity complete mRNA from the RNA of cells and tissues of animals, plants, insects, and eukaryotic microorganisms.
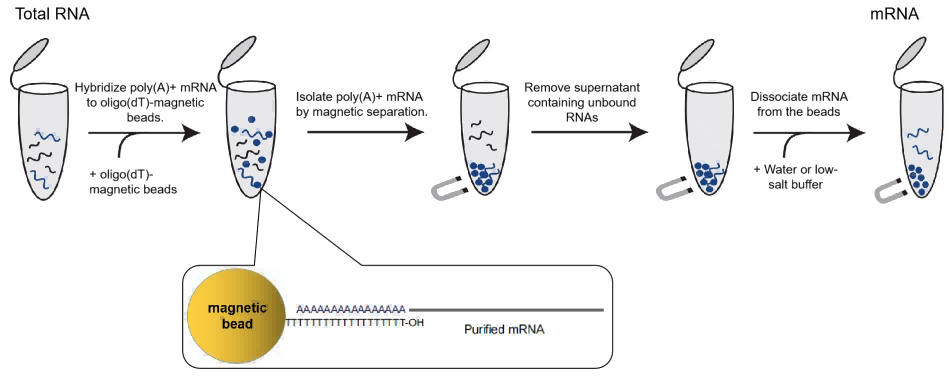
Figure 8. principle of enrichment and purification of mRNA magnetic beads
4.2 mRNA isolation beads performance
Hieff NGS™ mRNA Isolation Master Kit is independently developed by Yeasen for the isolation of mRNA from total RNA. The mRNA capture beads are micron-sized paramagnetic microspheres modified with oligo (dT). By combining with the mRNA with poly (A) tail, the mRNA is isolated and purified from 10 ng-4 μg total RNA with good integrity.
- High efficiency: mRNA purification can be completed within 45min
- High purity: oligo (dT) magnetic beads specifically bind mRNA
- Reliable: the obtained mRNA is suitable for NGS, in vitro translation, RT-PCR, and cDNA synthesis

Figure 9. mRNA isolation kit purification of mRNA from different RNA samples
Note: housekeeping gene yield represents the effect of mRNA recovery. rRNA related gene yield characterization rRNA removal efficiency
4.3 mRNA magnetic bead FAQ
(1) Why do magnetic beads agglomerate and how to solve it?
——Magnetic beads agglomeration will reduce the yield and purity of mRNA. The sample may contain many impurities, such as polysaccharides, proteins, and long-chain DNA. These impurities will lead to magnetic beads agglomeration. The solution is to blow the magnetic beads through a pipette. Genomic DNA can be removed by DNase treatment before purification. If the initial sample size is too large and exceeds the load of the magnetic beads, the magnetic beads will agglomerate too. It is recommended to operate according to the input amount in the manual.
(2) Why the mRNA yield is low?
——There are many reasons for low mRNA yield:
- The mRNA expression level of cells or tissues is low, so we can select the appropriate expression period samples or appropriately increase the sample total RNA input amount.
- The ratio of magnetic beads to samples is too low, which affects the combination of mRNA and magnetic beads. Try to increase the number of magnetic beads or reduce the sample input amount and volume.
- The hybridization time is too short, the incubation time can be increased to 10-15min;
- The elution is insufficient, it is necessary to appropriately increase the volume of the elution buffer, increase the elution time and temperature, or repeat the elution step twice.
(3) How to effectively eliminate rRNA?
——If the operation is improper, the rRNA will be non-specific bound to the magnetic beads along with the mRNA during the purification, especially when there is a large amount of total RNA input. Two rounds of binding are usually used in the purification process to effectively avoid rRNA pollution. Ensure that it is fully mixed during the washing experiment to remove the non-specific binding and try to remove rRNA pollution.
(4) For many downstream applications, is it necessary to elute mRNA, and will magnetic beads interfere with downstream enzyme reactions?
——Some downstream reactions can be carried out directly with magnetic beads without eluting mRNAs, such as mRNA fragmentation and reverse transcription in RNA library building. However, if PCR reactions are to be carried out, it is necessary to ensure that there is no genomic DNA pollution, otherwise many genomic amplification fragments will be generated, which will interfere with the experimental results. Detailed operations can be carried out according to the instructions of corresponding downstream reactions, such as RNA-Seq library construction
(5) Can the kit separate mRNA directly from cells or tissues?
——Not recommend! The lysate contains some components which will affect the binding of mRNA. It is recommended to use a special kit.
5. Yeasen magnetic beads product recommendation
Hieff NGS™ series magnetic beads as Yeasen star products have excellent performance and high production capacity to meet various applications, and seamlessly replace Beckman AMPure XP beads. Since its launch in 2018, Yeasen magnetic beads have won a good reputation in the industry, and sales have continued to rise. They are the best choice for good quality and low price.
Table 2. Yeasen magnetic beads product recommendation
| Product name | Cat# | Specification | Usage and application scenarios |
| Hieff NGS™ DNA Selection Beads | 12601ES03 | 1 mL | 👉NGS DNA purification and size selection 👉PCR product purification 👉Purification of enzyme digestion and ligation products |
| 12601ES08 | 5 mL | ||
| 12601ES56 | 60 mL | ||
| 12601ES75 | 450 mL | ||
| Hieff NGS™ RNA Cleaner | 12602ES03 | 1 mL | 👉RNA purification 👉Purification of total RNA samples after rRNA removal 👉Purification of RNA labeled products |
| 12602ES08 | 5 mL | ||
| 12602ES56 | 60 mL | ||
| 12602ES75 | 450 mL | ||
| Hieff NGS™ mRNA Isolation Master Kit | 12603ES24 | 24 T | 👉mRNA isolation and purification 👉In vitro transcription mRNA purification |
| 12603ES96 | 96 T |
Regarding Reading
About NGS-related technology, how much do you know?
5 minutes to understand the past and present of DNA selection beads (including result analysis and FAQ interpretation)
