Description
Eukaryotes mRNA forms a special structure at the 5'end after transcription, which is the cap structure. The cap structure plays an important role in the stability, transportation, and translation of mRNA. The vaccinia virus capping enzyme is an effective enzyme that can catalyze the formation of the cap structure. It"s composed of two subunits D1 and D12. It also has RNA triphosphatase activity, guanylate acyltransferase activity, and guanine methyltransferase activity that could connect the 7-methylguanine cap structure (m7Gppp) to the 5'end of the RNA (m7Gppp5'N). Vaccinia virus capping enzyme can cap the RNA in correct direction within one hour when present at a suitable concentration of capping buffer, guanosine triphosphate (GTP), S-adenosylmethionine (SAM), etc.
Feature
- Isolated from a recombinant source
- Tested for the absence of endonucleases, exonucleases, RNases
Application
- Capping mRNA before translation assays/in vitro translation
- Labeling 5´ end of mRNA
Specification
| Source | Recombinant E. coli with vaccinia virus capping enzyme gene |
| Optimum Temperature | 37℃ |
| Storage Buffer | 20 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM DTT, 0.1 mM EDTA, 0.1% Triton X-100, 50% glycerin |
| Unit Definition | 1 unit: The amount of enzyme required to incorporate 10 pmol GTP (α- 32P) into a transcript with 80 nucleotides (80 nt) at 37℃ within 1 hour. |
Components
| Components No. | Name | 10615ES84 (2,000 U) | 10615ES92 (10,000 U) | 10615ES96 (100,000 U) | 10615ES99 (5 MU) |
| 10615 | mRNA Vaccinia Capping Enzyme (10 U/μL) | 200 μL | 1 mL | 10 mL | 500 mL |
Shipping and Storage
mRNA Vaccinia Capping Enzyme GMP-grade products are shipped with dry ice and can be stored at -15℃ ~ -25℃ for one year.
Figures
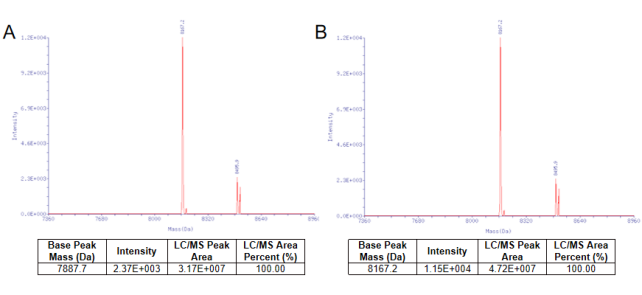
Figure 1. The capping efficiency almost reached 100% by the vaccinia capping system from YEASEN. Nucleic acid mass spectrometry before the cap reaction was shown in Figure 1A. Nucleic acid mass spectrometry after the cap reaction was shown in Figure 1B.
Payment & Security
Your payment information is processed securely. We do not store credit card details nor have access to your credit card information.
Inquiry
You may also like
FAQ
The product is for research purposes only and is not intended for therapeutic or diagnostic use in humans or animals. Products and content are protected by patents, trademarks, and copyrights owned by Yeasen Biotechnology. Trademark symbols indicate the country of origin, not necessarily registration in all regions.
Certain applications may require additional third-party intellectual property rights.
Yeasen is dedicated to ethical science, believing our research should address critical questions while ensuring safety and ethical standards.

