Description
The Hieff NGS™ OnePot Pro DNA Library Prep Kit V4 is a next-generation enzymatic library preparation kit designed for use with Illumina and MGI high-throughput sequencing platforms. Compared to traditional library preparation methods, this kit utilizes high-quality fragmentation enzymes, eliminating the need for cumbersome sonication processes. The fragmentation and end-repair modules are combined into a single step, significantly reducing both the time and cost of library preparation. This kit is suitable for a wide range of sample types, including plant and animal genomes, microbial genomes, and more, with a sample input range of 1 ng to 1 μg. The enzymatic fragmentation ensures uniform fragment sizes across different species, with minimal inter-species variation. Additionally, this kit can be paired with Illumina or MGI adapters and primers for sequencing on Illumina or MGI high-throughput platforms.
Specifications
|
Cat.No. |
12972ES08 / 12972ES24 / 12972ES96 |
|
Size |
8 T / 24 T / 96 T |
Components
|
Components No. |
Name |
12972ES08 |
12972ES24 |
12972ES96 |
|
12972-A |
80 μL |
240 μL |
960 μL |
|
|
12972-B |
Smearase™ Enzyme 4.0 |
80 μL |
240 μL |
960 μL |
|
12972-C |
Ligation Enhancer 4.0 |
240 μL |
720 μL |
3×960 μL |
|
12972-D |
Rapid DNA Ligase 4.0 |
80 μL |
240 μL |
2×480 μL |
|
12972-E |
2×Ultima HF Amplification Mix |
200 μL |
600 μL |
3×800 μL |
Storage
This product should be stored at -25~-15℃ for 1 year.
Notes
1. About the operation
1. Please operate with lab coats and disposable gloves,for your safety.
2. Thaw components at room temperature. After thawing, mix thoroughly by vortexing, spin the tube briefly and place them on ice for later use.
3. When preparing the reaction solution of each step, it is recommended to use a pipette to mix well or gently shake. Vigorous shaking may cause a decrease in library output.
4. It is highly recommended to use filtered pipet tips to avoid cross-contamination. Be sure to change pipet tips when processing different samples.
5. Improper operations may very likely cause aerosol contaminations, impacting the accuracy of result. Mandatory physical isolation of PCR reaction mixing regions and PCR product purification assay regions is recommended. Equipped with equipment such as specialized pipettes for library construction.Perform routine cleaning for each area by wiping the surfaces with 0.5% sodium hypochlorite or 10% bleach
6. This product is for research use only.
2. DNA Fragmentation1. The kit is compatible with 100 pg - 1000 ng of input DNA. It is highly recommended to use high-quality input DNA with A260/A280 = 1.8-2.0.
2. Following experiments could be impacted if high concentrations of salts like the metal chelating agent were introduced with the input DNA. We recommend eluting the DNA sample in ddH2O for fragmentation.
3. Please refer to table 6 for the fragmentation time of standard DNA samples. The kit has low fragmentation bias and provides uniform GC coverage for DNA samples with a wide range of GC compositions. Please adjust the fragmentation time based on your experimental requirements.
4. For accurate fragmentation, please prepare the reaction on ice.
3. Adapter Ligation
1. Illumina or MGI Long Adapter (Barcoded Adapter) kits and short Adapter kits are available for customers to choose according to their experimental requirements.
2. Selecting high-quality, commercial adapters was recommended. If self-made adapters are selected, please entrust a company with experience in NGS primer synthesis and remark the need for strict contamination control. In addition, it is recommended to prepare DNA annealing solution in a clean bench and only operate one type of adapter each time to prevent cross-contamination.
3. Please thaw the adapters on the ice or at 4°C; when operating at room temperature, the laboratory temperature should not exceed 25°C to prevent the adapters from denaturing.
4. The adapters’ quality and concentration will directly affect the ligation efficiency and the library yield. Too high concentration of adapters favors adapter dimer formation while too little adapter reduces ligation rate and library yield. Corresponding dilutions with TE Buffer according to Input DNA amount when using Adapter.Table 2 -3 lists the recommended Adapter dilution methods for different Input DNA amounts using this kit.
Table 1 The recommended Illumina adapter amount for different input DNA
|
Input DNA |
Adapter Dilution( Volume of Adapter : Total Volume) |
Concentration |
|
1 ng |
30-Fold (1 : 30) |
0.5 μM |
|
10 ng |
7.5-Fold (1 : 7.5) |
2 μM |
|
100 ng |
3-Fold (1 : 3) |
5 μM |
|
1000 ng |
1.5-Fold (1 : 1.5) |
10 μM |
Table 2 The recommended MGI adapter amount for different input DNA
|
Input DNA |
Adapter Dilution( Volume of Adapter : Total Volume) |
Concentration |
|
1 ng |
20-Fold (1 : 20) |
0.5 μM |
|
10 ng |
5-Fold (1 : 5) |
2 μM |
|
100 ng |
2-Fold (1 : 2) |
5 μM |
|
1000 ng |
undiluted |
10 μM |
4. Bead-based DNA Cleanup and Size Selection
1. The DNA fragment size selection step can be performed either after adapter ligation or after library amplification.
2. It is recommended to perform size-selection right after adapter ligation if the input DNA amount is more than 50 ng; otherwise, please perform size-selection after amplification.
3. The Ligation Enhancer contains a high concentration of PEG, which may cause a significant impact on accurate size-selection. Thus, if size-selection is to be performed right after adapter ligation, it is strongly recommended to add a beads clean-up step before the size-selection. Size selection step can be performed directly if it is performed before the end repair/dA-tailing or after the library amplification.
4. The magnetic beads should be equilibrated at room temperature prior to use, otherwise the yield will decrease and the size selecting effect will be affected.
5. The magnetic beads should be mixed well by vortex or pipetting prior to use.
6. Do not aspirate the beads when transferring the supernatant, even trace amounts of the beads may impact the following reactions.
7. The 80% ethanol should be freshly prepared, otherwise it will affect the recovery efficiency.
8. For accurate size-selection, it is recommended to start with a volume of more than 100 μL. If less, it is recommended to bring the volume up to 100 μL with ultra-pure water.
9. The magnetic beads should be dried at room temperature before eluting the product. Insufficient dryness will easily cause ethanol residual to affect subsequent reactions; excessive dryness will cause the magnetic beads to crack and reduce the purification yield. Normally, drying at room temperature for 3-5 minutes is enough to allow the beads to fully dry.
10. If needed, the purified or size-selected DNA samples eluted in 0.1× TE buffer can be stored at 4°C for 1-2 weeks or at -20°C for a month.
5. Library Amplification
1. Whether or not to perform library amplification depends on the amount of DNA input, types of the adapters, the sequencing data applications, etc. The amplification step is required if using partial adapters. When using full-length adapters, if the input DNA<200 ng, it is recommended to perform amplification; otherwise, amplification is not necessary.
2. Amplification cycle numbers should be strictly controlled. Insufficient amplification may lead to low library yield; Over-amplification may introduce increased bias, errors, duplicated read, and chimeric products. Table 3 lists recommended cycle numbers targeting the library yield of 1 μg.
Table 3 The recommended number of cycles to generate 1,000 ng of library yield
|
Input DNA |
Number of cycles required to generate 1 μg of library yield |
|
1000 ng |
2 - 4 |
|
500 ng |
2 - 4 |
|
250 ng |
4 - 6 |
|
100 ng |
5 - 7 |
|
50 ng |
7 - 9 |
|
1 ng |
12 - 14 |
Note:
1.Table 3 shows the number of loop parameters using high-quality Input DNA tests of around 200 bp.The FFPE DNA quality varies greatly, and when the DNA quality is poor or the library length is long, the number of cycles needs to be appropriately increased to obtain sufficient libraries.
2.If size selection is required during the library building process, higher cycle number for Library Amplification is recommended; otherwise, lower cycle number is recommended.
3.If incomplete adapters are used, at least 2 cycles need to be amplified to form a complete adapter.
6. Library Quality Analysis
1. The constructed libraries quality is generally analyzed by measuring the concentrations and size distributions.
2. Libraries’concentrations can be measured by fluorescent-based methods such as Qubit and PicoGreen or qPCR.
3. It is NOT recommended to use absorbance-based quantification methods such as NanoDrop.
4. It is recommended to use qPCR method for library quantification: fluorescent-based methods such as Qubit and PicoGreen cannot differentiate the incomplete dsDNA structures (inserts with no adapter or with only one of the ends ligated with adapter) from the complete libraries. The qPCR method will only amplify and measure the complete libraries with both ends ligated with adapters (the sequencable libraries), thus providing a more accurate measurement for loading.
5. The size distribution of libraries can be analyzed using Agilent Bioanalyzer or other devices based on the principles of capillary electrophoresis or microfluidics.
7. Other Materials
1. DNA purification magnetic beads: Hieff NGSTM DNA Selection Beads (Yeasen Cat#12601) or AMPure® XP Beads (A63880) or other equivalent products.
2. Adapters: Complete Adapter for Illumina: Yeasen Cat#13519-13520; 384 Dual CDI Primers: Yeasen Cat#12412~Cat#12413; 384 Unique Dual Index (UDI) Primers: Yeasen Cat#12327~Cat#13330; UMI UDI Adapters: Yeasen Cat#13370~Cat#13371; Complete Adapter for MGI: Yeasen Cat#13360-13362. DNA Primer Mix:Cat#12190 or Cat#12191.
3. Library quality analysis: Agilent 2100 Bioanalyzer DNA 1000 Chip/ High Sensitivity Chip or other equivalent products; library quantitative reagents.
4. Other materials: absolute ethanol, sterile ultrapure water, low retention pipette tips, PCR tube, magnetic stands, thermal cycler, etc.
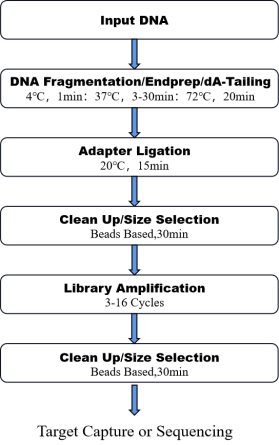
8. Workflow
Figure 1. The workflow of OnePot Pro DNA Library Prep Kit
Payment & Security
Your payment information is processed securely. We do not store credit card details nor have access to your credit card information.
Inquiry
You may also like
FAQ
The product is for research purposes only and is not intended for therapeutic or diagnostic use in humans or animals. Products and content are protected by patents, trademarks, and copyrights owned by Yeasen Biotechnology. Trademark symbols indicate the country of origin, not necessarily registration in all regions.
Certain applications may require additional third-party intellectual property rights.
Yeasen is dedicated to ethical science, believing our research should address critical questions while ensuring safety and ethical standards.