Description
Cell Counting Kit-8 (CCK-8) is a fast and highly sensitive detection kit based on the WST-8 (2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium monosodium salt) reagent. WST-8 is an upgraded reagent of MTT and can be reduced to a highly water-soluble orange-yellow formazan by dehydrogenases in the mitochondria. The amount of the formazan generated is proportional to the number of living cells. The OD value of formazan at 450nm detected by a microplate reader can indirectly indicate the number of viable cells. This kit is widely used for drug screening, cell proliferation and cytotoxicity assays, tumor drug sensitivity testing, and activity detection of biological factors.
Advantages of CCK-8 method
1. Comparison of the CCK-8 method with other cell proliferation/toxicity detection methods.
|
Detection method |
MTT method |
XTT method |
WST - 1 method |
CCK 8 method |
|---|---|---|---|---|
|
Water solubility of the formazan product |
Low (need to add organic solvent to dissolve and then test) |
High |
High |
High |
|
Product characteristics |
Powder |
2 bottles of solution |
Solution |
1 bottle of solution |
|
Method of use |
Mix into solution and use |
The solution was prepared just before use. |
Out of the box |
Out of the box |
|
Detection sensitivity |
High |
Very high |
Very high |
High |
|
Detection time |
Long |
Short |
Short |
The shortest |
|
Detection wavelength |
560-600 nm |
420-480 nm |
420-480 nm |
430-490 nm |
|
cytotoxicity |
High toxicity, complete disappearance of cell morphology |
Low toxicity, cell morphology unchanged |
Low toxicity, cell morphology unchanged |
Low toxicity, cell morphology unchanged |
|
Reagent stability |
General |
Low |
General |
High |
|
Bulk sample testing |
Feasible |
Appropriate |
Appropriate |
Appropriate |
2. Phenol red and serum do not interfere with CCK-8 detection.
3. As the cytotoxicity of CCK-8 reagent is very low, the optimal measurement time can be determined by repeatedly reading with a microplate reader at different times after adding CCK-8 reagent.
Shipping and Storage
The product could be stored at -25~-15ºC in a dry and dark place for two years.
Precautions
1. In the first experiment, it is recommended to determine the optimal number of cells plated and the optimal incubation time of CCK-8 reagent.
2. If possible, use a multi-channel pipette to reduce variations between replicate wells. Avoid bubbles in the experiment as bubbles will interfere with the OD reading. When adding the CCK-8 reagent, it is recommended to add it close to the wall of the culture plate rather than inserting it into the culture medium.
3. White blood cells may require a longer incubation time.
4. When using a standard 96-well plate, the number of cells plated is at least 1,000 cells/well (100 μL). The detection sensitivity for white blood cells is relatively low, so it is recommended to plate at least 2,500 cells/well (100 μL). If using a 24-well or 6-well plate, calculate the corresponding number of cells for each well and add the appropriate volume of CCK-8 reagent (the volume of CCK-8 reagent added is 10% of the volume of the culture medium in each well of the plate).
5. If the 450 nm filter is not available, a filter with absorbance between 430-490 nm can be used, but the 450 nm filter can achieve the highest detection sensitivity.
6. Phenol red does not affect the measurement as the absorbance of phenol red in the medium can be eliminated by subtracting the absorbance of the blank group.
7. For your safety and health, please wear the lab coat and disposable gloves when operating the experiment.
Instructions
I. Make a standard curve
1. Prepare the cell suspension, determine the cell density, and then plate cells.
2. Sequentially dilute the cells with the culture medium according to a certain ratio (such as 1:2 ratio) to form a cell concentration gradient, generally 3-5 cell concentration gradients, 3-6 replicate wells for each concentration.
3. After plated, culture the cells for 2-4 hours to make them adhesion. Then, add the CCK-8 reagent, incubate for a certain period of time and measure the OD value. Plot a standard curve with the cell number as the X-axis and the OD value as the Y-axis. According to this standard curve, the cell number in an unknown sample can be determined (The premise of using this standard curve is that the experimental conditions should be consistent).
II. Cell viability assay
1. Plate cells (100 μL/well) in a 96-well plate. Place the plate in an incubator (37°C, 5% CO2) for a period of time for pre-incubation.
2. Add 10 μL of CCK-8 reagent to each well (be careful not to introduce bubbles to the wells, since they interfere with the OD reading) and mix gently.
3. Place the plate in the incubator and incubate for 1-4 hours.
4. Measure the absorbance at 450 nm with a microplate reader.
5. If the OD value is not measured immediately, add 10 μL of 0.1 M HCl solution or 1% w/v SDS solution to each well, cover the plate and store it with protection from light at room temperature. The absorbance value will not change within 24 hours.
III. Cell proliferation and cytotoxicity assay
1. Plate cells (100 μL/well) in a 96-well plate. Place the plate in an incubator (37°C, 5% CO2) for 24 hours for pre-incubation.
2. Add 10 μL of different concentrations of the substance to be tested to the plate.
3. Place the plate in the incubator and incubate for a certain period of time (such as 6, 12, 24, or 48 hours).
4. Add 10 μL of CCK-8 reagent to each well (be careful not to introduce bubbles to the wells, since they interfere with the OD reading) and mix gently.
5. Place the plate in the incubator and incubate for 1-4 hours.
6. Measure the absorbance at 450 nm with a microplate reader.
7. If the OD value is not measured immediately, add 10 μL of 0.1 M HCl solution or 1% w/v SDS solution to each well, cover the plate and store it with protection from light at room temperature. The absorbance value will not change within 24 hours.
Note: If the substance is oxidizing or reducing, replace the old culture medium with fresh medium (remove the old medium, wash the cells twice with fresh medium, and then add fresh medium) before adding the CCK-8 reagent to eliminate the effect of the substance. If the substance itself has only a slight effect on the absorbance value, there is no need to replace the culture medium and the effect of the substance can be eliminated by subtracting the absorptance value of a blank group with the substance.
Calculation formula
Cell survival rate =[(A-C) /(B-C)] x100%
Inhibition rate =[(B-A) /(B-C)] x100%
A: The absorbance of the experimental group (the absorbance of the culture medium containing cells, CCK-8 reagent, and the substance to be tested)
B: The absorbance of the control group (the absorbance of the culture medium containing cells, CCK-8 reagent)
C: The absorbance of blank group (the absorbance of the culture medium containing CCK-8 reagent)
Citations

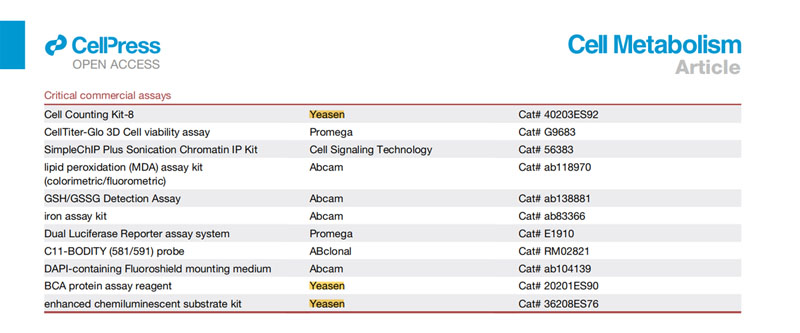
[1] Sun L, Li P, Ju X, et al. In vivo structural characterization of the SARS-CoV-2 RNA genome identifies host proteins vulnerable to repurposed drugs. Cell. 2021;184(7):1865-1883.e20. doi:10.1016/j.cell.2021.02.008(IF:41.584)
[2] Wei S, Zhao Q, Zheng K, et al. GFAT1-linked TAB1 glutamylation sustains p38 MAPK activation and promotes lung cancer cell survival under glucose starvation. Cell Discov. 2022;8(1):77. Published 2022 Aug 9. doi:10.1038/s41421-022-00423-0(IF:38.079)
[3] Chen X, Zhang D, Su N, et al. Visualizing RNA dynamics in live cells with bright and stable fluorescent RNAs. Nat Biotechnol. 2019;37(11):1287-1293. doi:10.1038/s41587-019-0249-1(IF:31.864)
[4] Yang F, Xiao Y, Ding JH, et al. Ferroptosis heterogeneity in triple-negative breast cancer reveals an innovative immunotherapy combination strategy [published online ahead of print, 2022 Oct 11]. Cell Metab. 2022;S1550-4131(22)00411-9. doi:10.1016/j.cmet.2022.09.021(IF:31.373)
[5] Rong QX, Wang F, Guo ZX, et al. GM-CSF mediates immune evasion via upregulation of PD-L1 expression in extranodal natural killer/T cell lymphoma. Mol Cancer. 2021;20(1):80. Published 2021 May 29. doi:10.1186/s12943-021-01374-y(IF:27.401)
[6] Xia B, Shen X, He Y, et al. SARS-CoV-2 envelope protein causes acute respiratory distress syndrome (ARDS)-like pathological damages and constitutes an antiviral target. Cell Res. 2021;31(8):847-860. doi:10.1038/s41422-021-00519-4(IF:25.617)
[7] Yang X, Zhao X, Zhu Y, et al. FBXO34 promotes latent HIV-1 activation by post-transcriptional modulation. Emerg Microbes Infect. 2022;11(1):2785-2799. doi:10.1080/22221751.2022.2140605(IF:19.568)
[8] Zhou Z, Zhang X, Lei X, et al. Sensing of cytoplasmic chromatin by cGAS activates innate immune response in SARS-CoV-2 infection. Signal Transduct Target Ther. 2021;6(1):382. Published 2021 Nov 3. doi:10.1038/s41392-021-00800-3(IF:18.187)
[9] Li M, Hao B, Zhang M, et al. Melatonin enhances radiofrequency-induced NK antitumor immunity, causing cancer metabolism reprogramming and inhibition of multiple pulmonary tumor development. Signal Transduct Target Ther. 2021;6(1):330. Published 2021 Sep 1. doi:10.1038/s41392-021-00745-7(IF:18.187)
[10] Qi S, Zhu Y, Liu X, et al. WWC proteins mediate LATS1/2 activation by Hippo kinases and imply a tumor suppression strategy. Mol Cell. 2022;82(10):1850-1864.e7. doi:10.1016/j.molcel.2022.03.027(IF:17.970)
[11] Zhu J, Li X, Cai X, et al. Arginine monomethylation by PRMT7 controls MAVS-mediated antiviral innate immunity. Mol Cell. 2021;81(15):3171-3186.e8. doi:10.1016/j.molcel.2021.06.004(IF:17.970)
[12] Teng KX, Niu LY, Xie N, Yang QZ. Supramolecular photodynamic agents for simultaneous oxidation of NADH and generation of superoxide radical. Nat Commun. 2022;13(1):6179. Published 2022 Oct 19. doi:10.1038/s41467-022-33924-3(IF:17.694)
[13] Zhong J, Guo Y, Lu S, et al. Rational design of a sensitivity-enhanced tracer for discovering efficient APC-Asef inhibitors. Nat Commun. 2022;13(1):4961. Published 2022 Aug 24. doi:10.1038/s41467-022-32612-6(IF:17.694)
[14] Liu F, Wang X, Duan J, et al. A Temporal PROTAC Cocktail-Mediated Sequential Degradation of AURKA Abrogates Acute Myeloid Leukemia Stem Cells. Adv Sci (Weinh). 2022;9(22):e2104823. doi:10.1002/advs.202104823(IF:16.806)
[15] Ji C, Qiu M, Ruan H, et al. Transcriptome Analysis Revealed the Symbiosis Niche of 3D Scaffolds to Accelerate Bone Defect Healing. Adv Sci (Weinh). 2022;9(8):e2105194. doi:10.1002/advs.202105194(IF:16.806)
[16] Feng L, Dou C, Xia Y, et al. Neutrophil-like Cell-Membrane-Coated Nanozyme Therapy for Ischemic Brain Damage and Long-Term Neurological Functional Recovery. ACS Nano. 2021;15(2):2263-2280. doi:10.1021/acsnano.0c07973(IF:15.881)
[17] Wang Z, Gong X, Li J, et al. Oxygen-Delivering Polyfluorocarbon Nanovehicles Improve Tumor Oxygenation and Potentiate Photodynamic-Mediated Antitumor Immunity. ACS Nano. 2021;15(3):5405-5419. doi:10.1021/acsnano.1c00033(IF:15.881)
[18] Jiang Z, He L, Yu X, et al. Antiangiogenesis Combined with Inhibition of the Hypoxia Pathway Facilitates Low-Dose, X-ray-Induced Photodynamic Therapy [published online ahead of print, 2021 Jun 25]. ACS Nano. 2021;10.1021/acsnano.1c01063. doi:10.1021/acsnano.1c01063(IF:15.881)
[19] Gong X, Li J, Xu X, et al. Microvesicle-inspired oxygen-delivering nanosystem potentiates radiotherapy-mediated modulation of tumor stroma and antitumor immunity. Biomaterials. 2022;290:121855. doi:10.1016/j.biomaterials.2022.121855(IF:15.304)
[20] Deng J, Xu W, Lei S, et al. Activated Natural Killer Cells-Dependent Dendritic Cells Recruitment and Maturation by Responsive Nanogels for Targeting Pancreatic Cancer Immunotherapy. Small. 2022;18(44):e2203114. doi:10.1002/smll.202203114(IF:15.153)
Payment & Security
Your payment information is processed securely. We do not store credit card details nor have access to your credit card information.
Inquiry
You may also like
FAQ
The product is for research purposes only and is not intended for therapeutic or diagnostic use in humans or animals. Products and content are protected by patents, trademarks, and copyrights owned by Yeasen Biotechnology. Trademark symbols indicate the country of origin, not necessarily registration in all regions.
Certain applications may require additional third-party intellectual property rights.
Yeasen is dedicated to ethical science, believing our research should address critical questions while ensuring safety and ethical standards.