Description
GMyc-PCR Mycoplasma Detection Kit mainly uses the PCR method to detect Mycoplasma infection of various biological materials (such as cell culture, experimental animal secretions, animal serum, etc.). It combines several advantages: sensitive, specific, rapid, and can be detected directly with cell culture supernatants. This product detects mycoplasma in biological materials such as cultured cells by PCR method. The primers used are designed according to the conserved region of the 16S-23S rRNA sequence of mycoplasma, and only specifically amplify the mycoplasma DNA, with high detection sensitivity and specificity. PCR amplification and electrophoresis analysis only take a few hours, and the operation is convenient and simple.
Cell culture is a common experiment in life science research. Unlike other commonly used experimental methods, cell culture is a dynamic continuous process, and cells often respond to manipulation errors or contaminants that often exhibit abnormal cell states or medium appearance. If it is contaminated by mycoplasma, the cell morphology has no obvious change, and it is easy to be overlooked. It is often not found until the pollution is very serious. There may be hundreds of mycoplasmas on the contaminated cell membrane, these mycoplasmas compete for nutrients and release toxic metabolites, seriously affecting the experimental results.
Studies have shown that at least 20 kinds of mycoplasma can contaminate cells, among which the most common are: oral Mycoplasma (M. orale), Mycoplasma arginine (M. arginini), Mycoplasma hyorhinis (M. hyorhinis), Mycoplasma fermentum (M. fermentans), Mycoplasma hominis (M. hominis), Mycoplasma salivarius (M. salivarium), Mycoplasma pulmonary (M. pulmonis) and Mycoplasma pear (M. pirum). The mycoplasma contamination rate of cultured cells ranges from 4% to 92%. The sources of contamination include the working environment, the operator itself (some mycoplasmas are normal flora of the human body), culture medium, serum, cell cross-contamination, experimental equipment, and used Contamination of the original tissue or organ from which cells were prepared.
Identifying the underlying cause of problems during cell culture is a difficult and time-consuming task, where any sudden changes should be suspected, and good testing practices and regular testing for mycoplasma contamination are necessary. There are many methods for the detection of mycoplasma, such as direct culture, DNA fluorescence staining, ELISA, and PCR methods.
Yeasen offers comprehensive solutions for Mycoplasma contamination. Explore our related products: Concept of Mycoplasma and the Impact of Contamination
Feature
- The primers used were designed according to the conserved region of Mycoplasma 16S-23S rRNA sequence
- Only amplified mycoplasma DNA specifically
- Excellent sensitivity and specificity
- More than 20 mycoplasma species could be detected
Application
- Mycoplasma Detection
Components
| Components No. | Name | 40601ES10 (10 assays) | 40601ES20 (20 assays) |
| 40601-A | GMyc-1st PCR Mix | 250 µL | 2×250 µL |
| 40601-B | GMyc-2nd PCR Mix | 250 µL | 2×250 µL |
| 40601-C | Positive Control Template N | 20 µL | 20 µL |
[Notes] 1. When not in use for a long time, it can be stored frozen at -85~-65℃.
2. The PCR reaction is extremely sensitive. In order to prevent false positives, a positive control is added at the end when adding samples.
Storage
This product could be stored at -25~-15℃ for 18 months. If it is not used for a long time, please keep it away from light.
Figures
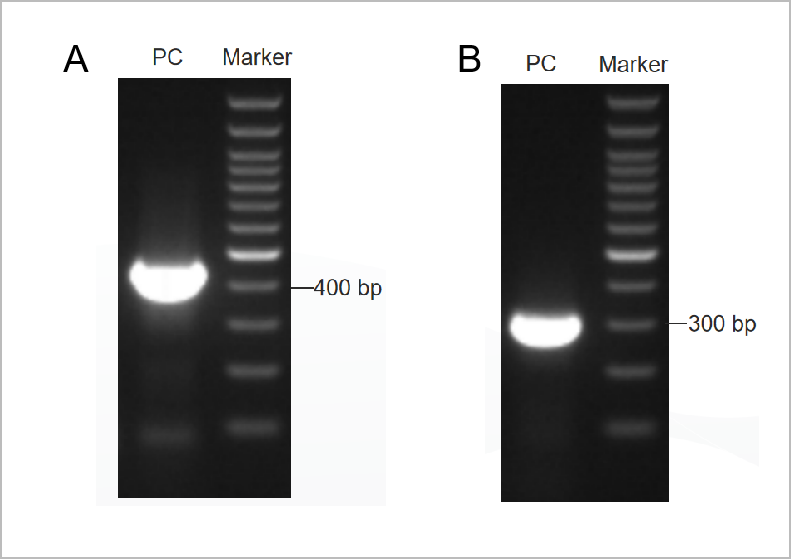
Figure 1. The electrophoretogram result of the PCR Mycoplama Test Kit.
The first round electrophoretogram was shown in Figure 1A and the second round electrophoretogram was shown in Figure 1B.(M: 1kb marker, 1-2: positive controls, 3: the negative control)
[1] Rao XS, Cong XX, Gao XK, et al. AMPK-mediated phosphorylation enhances the auto-inhibition of TBC1D17 to promote Rab5-dependent glucose uptake. Cell Death Differ. 2021;28(12):3214-3234. doi:10.1038/s41418-021-00809-9(IF:15.828)
[2] Guo F, Li L, Li J, et al. Single-cell multi-omics sequencing of mouse early embryos and embryonic stem cells. Cell Res. 2017;27(8):967-988. doi:10.1038/cr.2017.82(IF:15.606)
[3] Hao Y, He B, Wu L, et al. Nuclear translocation of p85β promotes tumorigenesis of PIK3CA helical domain mutant cancer. Nat Commun. 2022;13(1):1974. Published 2022 Apr 13. doi:10.1038/s41467-022-29585-x(IF:14.919)
[4] Shu X, Liu M, Lu Z, et al. Genome-wide mapping reveals that deoxyuridine is enriched in the human centromeric DNA. Nat Chem Biol. 2018;14(7):680-687. doi:10.1038/s41589-018-0065-9(IF:13.843)
[5] Li X, Xiong X, Wang K, et al. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat Chem Biol. 2016;12(5):311-316. doi:10.1038/nchembio.2040(IF:12.709)
[6] Sun L, Yang X, Huang X, et al. 2-Hydroxylation of Fatty Acids Represses Colorectal Tumorigenesis and Metastasis via the YAP Transcriptional Axis. Cancer Res. 2021;81(2):289-302. doi:10.1158/0008-5472.CAN-20-1517(IF:12.701)
[7] Sun Z, Zhang Z, Wang QQ, Liu JL. Combined Inactivation of CTPS1 and ATR Is Synthetically Lethal to MYC-Overexpressing Cancer Cells. Cancer Res. 2022;82(6):1013-1024. doi:10.1158/0008-5472.CAN-21-1707(IF:12.701)
[8] Song J, Zhuang Y, Zhu C, et al. Differential roles of human PUS10 in miRNA processing and tRNA pseudouridylation. Nat Chem Biol. 2020;16(2):160-169. doi:10.1038/s41589-019-0420-5(IF:12.154)
[9] He B, Pan H, Zheng F, et al. Long noncoding RNA LINC00930 promotes PFKFB3-mediated tumor glycolysis and cell proliferation in nasopharyngeal carcinoma. J Exp Clin Cancer Res. 2022;41(1):77. Published 2022 Feb 24. doi:10.1186/s13046-022-02282-9(IF:11.161)
[10] Tang B, Liu BH, Liu ZY, Luo MY, Shi XH, Pang DW. Quantum Dots with a Compact Amphiphilic Zwitterionic Coating. ACS Appl Mater Interfaces. 2022;14(24):28097-28104. doi:10.1021/acsami.2c04438(IF:9.229)
[11] Huang C, Zhang Z, Chen L, et al. Acetylation within the N- and C-Terminal Domains of Src Regulates Distinct Roles of STAT3-Mediated Tumorigenesis. Cancer Res. 2018;78(11):2825-2838. doi:10.1158/0008-5472.CAN-17-2314(IF:9.130)
[12] Wu X, Yu M, Zhang Z, et al. DDB2 regulates DNA replication through PCNA-independent degradation of CDT2. Cell Biosci. 2021;11(1):34. Published 2021 Feb 8. doi:10.1186/s13578-021-00540-5(IF:7.133)
[13] Wang J, Zhang Y, Liu X, Liu H. Optimizing Adaptive Therapy Based on the Reachability to Tumor Resistant Subpopulation. Cancers (Basel). 2021;13(21):5262. Published 2021 Oct 20. doi:10.3390/cancers13215262(IF:6.639)
[14] Feng W, Liu R, Xie X, et al. SUMOylation of α-tubulin is a novel modification regulating microtubule dynamics. J Mol Cell Biol. 2021;13(2):91-103. doi:10.1093/jmcb/mjaa076(IF:6.216)
[15] Yu M, Hu X, Yan J, Wang Y, Lu F, Chang J. RIOK2 Inhibitor NSC139021 Exerts Anti-Tumor Effects on Glioblastoma via Inducing Skp2-Mediated Cell Cycle Arrest and Apoptosis. Biomedicines. 2021;9(9):1244. Published 2021 Sep 17. doi:10.3390/biomedicines9091244(IF:6.081)
[16] Ren S, Cai Y, Hu S, et al. Berberine exerts anti-tumor activity in diffuse large B-cell lymphoma by modulating c-myc/CD47 axis. Biochem Pharmacol. 2021;188:114576. doi:10.1016/j.bcp.2021.114576(IF:5.858)
[17] Wen F, Sun X, Sun C, et al. TAGLN Is Downregulated by TRAF6-Mediated Proteasomal Degradation in Prostate Cancer Cells. Mol Cancer Res. 2021;19(7):1113-1122. doi:10.1158/1541-7786.MCR-20-0513(IF:5.852)
[18] Tang B, Sun EZ, Zhang ZL, et al. Sphingomyelin-Sequestered Cholesterol Domain Recruits Formin-Binding Protein 17 for Constricting Clathrin-Coated Pits in Influenza Virus Entry. J Virol. 2022;96(5):e0181321. doi:10.1128/JVI.01813-21(IF:5.103)
[19] Hu J, Ren W, Qiu W, et al. Generation of induced pluripotent stem cell line (XDCMHi001-A) from an Ankylosing spondylitis patient with JAK2 mutation. Stem Cell Res. 2020;45:101788. doi:10.1016/j.scr.2020.101788(IF:4.495)
[20] Xiao S, Yao X, Ye J, Tian X, Yin Z, Zhou L. Epigenetic modification facilitates proline synthase PYCR1 aberrant expression in gastric cancer [published online ahead of print, 2022 May 30]. Biochim Biophys Acta Gene Regul Mech. 2022;1865(6):194829. doi:10.1016/j.bbagrm.2022.194829(IF:4.490)
[21] Wang J, Zhang Y, Liu X, Liu H. Is the Fixed Periodic Treatment Effective for the Tumor System without Complete Information?. Cancer Manag Res. 2021;13:8915-8928. Published 2021 Nov 30. doi:10.2147/CMAR.S339787(IF:3.989)
[22] Yang X, Ren S, Rehman ZU, et al. Molecular characterization, expression, and functional identification of TANK-binding kinase 1 (TBK1) of the cow (Bos taurus) and goat (Capra hircus). Dev Comp Immunol. 2022;133:104444. doi:10.1016/j.dci.2022.104444(IF:3.636)
[23] Zheng D, Chang X, Liu Y, et al. 2-Methoxy-5((3,4,5-trimethosyphenyl)seleninyl) phenol reverses EGF-induced cell migration and invasion through down-regulation of MDM2 in breast cancer cell lines. Cancer Biol Ther. 2019;20(4):513-523. doi:10.1080/15384047.2018.1537578(IF:3.373)
[24] Xu F, Zhang S, Liu Z, et al. TEX9 and eIF3b functionally synergize to promote the progression of esophageal squamous cell carcinoma. BMC Cancer. 2019;19(1):875. Published 2019 Sep 3. doi:10.1186/s12885-019-6071-9(IF:2.933)
[25] Pan H, Sun L, Wang W, et al. Serum long non-coding RNA LOC553103 as non-specific diagnostic and prognostic biomarker for common types of human cancer. Clin Chim Acta. 2020;508:69-76. doi:10.1016/j.cca.2020.05.017(IF:2.615)
[26] Li N, Lin SM, Li Y, Sun J, Zhang L, Chen M. An induced pluripotent stem cell line (GZHMCi004-A) derived from a fetus with heterozygous G380R mutation in FGFR3 gene causing achondroplasia. Stem Cell Res. 2021;53:102322. doi:10.1016/j.scr.2021.102322(IF:2.020)
[27] Luo Q, Wei C, Long Y, et al. Generation of an ELTD1 knockout human embryonic stem cell line by the iCRISPR/Cas9 system. Stem Cell Res. 2021;53:102350. doi:10.1016/j.scr.2021.102350(IF:2.020)
[28] Liu YQ, Ling TW, Wang HY, Yang YH, Song WJ, Wang TC. Generation of an integration-free induced pluripotent stem cell line (LZUSHI001-A) from an epileptic patient with DGKG mutation. Stem Cell Res. 2022;61:102768. doi:10.1016/j.scr.2022.102768(IF:2.020)
[29] Chen M, Lin SM, Li N, Li Y, Li Y, Zhang L. An induced pluripotent stem cell line (GZHMCi003-A) derived from a fetus with exon 3 heterozygous deletion in RUNX2 gene causing cleidocranial dysplasia. Stem Cell Res. 2021;51:102166. doi:10.1016/j.scr.2021.102166(IF:2.020)
[30] Xu Y, Wang X, Qiu T, et al. Generation of an induced pluripotent stem cell line (FDCHI007-A) derived from a patient with developmental and epileptic encephalopathy Type 31 carrying heterozygous c.545C > A mutation in DNM1 gene. Stem Cell Res. 2022;60:102709. doi:10.1016/j.scr.2022.102709(IF:2.020)
[31] Fan T, He J, Wang Y, Yu J, Sun W. Generation of an induced pluripotent stem cell line (FDCHi006-A) from a 7-year-old girl with central precocious puberty. Stem Cell Res. 2021;56:102542. doi:10.1016/j.scr.2021.102542(IF:2.020)
[32] Gong X, Zheng Z, Yang T, Zheng H, Xiao X, Jia N. Generation of an isogenic gene-corrected iPSC line (OGHFUi001-A-1) from a type 1 early infantile epileptic encephalopathy (EIEE1) patient with a hemizygous R330L mutation in the ARX gene. Stem Cell Res. 2022;60:102693. doi:10.1016/j.scr.2022.102693(IF:2.020)
[33] Jia N, Gong X, Chen J, et al. Generation of an induced pluripotent stem cell line (OGHFUi001-A) from a type 1 early infantile epileptic encephalopathy with ARX mutation. Stem Cell Res. 2021;53:102367. doi:10.1016/j.scr.2021.102367(IF:2.020)
[34] Zhu W, Zhou Y, Wang Q, et al. Generation of a human induced pluripotent stem cell (iPSC) line from skin fibroblasts of a patient carrying an E363Q mutation in PSEN1 gene. Stem Cell Res. 2022;61:102769. doi:10.1016/j.scr.2022.102769(IF:2.020)
[35] Luo F, Long K, Li X, et al. Deficient of LRRC8A attenuates hypoxia-induced necrosis in 3T3-L1 cells. Biosci Biotechnol Biochem. 2020;84(6):1139-1145. doi:10.1080/09168451.2020.1730689(IF:1.516)
Payment & Security
Your payment information is processed securely. We do not store credit card details nor have access to your credit card information.
Inquiry
You may also like
FAQ
The product is for research purposes only and is not intended for therapeutic or diagnostic use in humans or animals. Products and content are protected by patents, trademarks, and copyrights owned by Yeasen Biotechnology. Trademark symbols indicate the country of origin, not necessarily registration in all regions.
Certain applications may require additional third-party intellectual property rights.
Yeasen is dedicated to ethical science, believing our research should address critical questions while ensuring safety and ethical standards.

