In recent years, the global incidence of infectious diseases has been on the rise, with pathogens becoming increasingly diverse and complex. A significant challenge in this scenario is that roughly half of the patients face the predicament of having unknown pathogens, making the rapid diagnosis of these pathogens a daunting task. Currently, clinical pathogen detection relies mainly on traditional culture methods, PCR techniques, metagenomic sequencing (mNGS), and pathogen-targeted sequencing (tNGS).
Real-time fluorescent quantitative PCR has gained prominence in the nucleic acid detection of SARS-CoV-2. Similarly, mNGS has garnered attention for its contributions to the fight against SARS-CoV-2 and has transitioned from clinical settings to everyday use. tNGS, offering the advantages of both PCR and NGS, such as speed, accuracy, and cost-effectiveness in sequencing services, is gaining increasing traction in the field of clinical testing.
Concurrently, the application of biological products has expanded into biomedicine, biological agriculture, bioenergy, biological manufacturing, environmental protection, and other domains. As the market share of biological products continues to grow, government bodies and relevant authorities have established standardized quality management systems and standards for these products. A particular area of focus pertains to the presence of nucleic acid residues from host cells in biological products.
Biological products like recombinant protein drugs, antibody drugs, vaccines, and cell and gene therapies are produced using consecutive strains or cell lines, potentially resulting in the retention of host nucleic acids in the final products. Residual host nucleic acids can have adverse consequences, such as uncontrolled cell proliferation leading to tumor formation or exacerbating immune responses by introducing viral genes. Hence, effective nucleic acid residue removal and rigorous residue detection are crucial for ensuring biological product safety and meeting research requirements. In the industry, qPCR/RT-qPCR methods are widely employed to develop detection kits for host nucleic acid residue detection in biological products.
Furthermore, commercial molecular enzymes are typically expressed using recombinant engineering strains, such as E. coli, resulting in the presence of host genomic DNA in these molecular enzymes. Additionally, environmental and human factors can introduce contaminated DNA into molecular enzyme products.
During the pathogen detection process, background bacterial nucleic acids from contamination may overshadow target nucleic acids with low abundance or be detected alongside target nucleic acids. This can impact the sensitivity of target detection or lead to false positive results, complicating the diagnosis and treatment decisions made by medical professionals.
In the development and production of quality control products for host nucleic acid residue testing, the presence of residual host nucleic acids, including those from humans, mice, E. coli, yeast, and others, in molecular enzymes can lead to inaccuracies in the quantification of host nucleic acid residue quality control products. This poses potential safety risks in the manufacturing of biological products.
YEASEN UCF.METM ultra-low residue molecular enzyme solution
In order to solve the problem of background bacteria and host nucleic acid residue interference, YEASEN has established a complete R&D and production platform for UCF.METM ultra-low residue molecular enzymes. And has achieved large-scale production of a variety of UCF.METM ultra-low residue molecular enzymes through material selection, environmental control, process optimization, and quality assurance.
YEASEN UCF.METM ultra-low residue molecular enzyme products
YEASEN reformed a complete set of molecular enzymes such as qPCR/RT-qPCR, NGS library building. At the same time, UCF.ME™ ultra-low residue process is used to process molecular enzyme products, we can provide a full set of high-performance and ultra-low host residue molecular enzyme raw materials for qPCR/RT-qPCR and NGS to improve the detection accuracy.
Table 1 The list of YEASEN UCF.METM ultra-low residue molecular enzyme products
|
Product Classification |
Product Name |
Catalog No. |
Criteria of E. coli gDNA quality control |
|
UCF.METM ultra-low residue enzyme products for qPCR/RT-qPCR |
Hieff UCF.METM Hotstart Sensitive Taq DNA Polymerase (5 U/μL) |
14314ES |
<0.005 copies/U |
|
Hifair UCF.METM V Reverse Transcriptase (200 U/μL) |
14608ES |
<0.005 copies/U |
|
|
UCF.METM Murine RNase Inhibitor (40 U/μL) |
14672ES |
<0.001 copies/U |
|
|
UCF.METM High Affinity RNase Inhibitor (40 U/μL) |
14675ES |
<0.001 copies/U |
|
|
UCF.METM Uracil DNA Glycosylase(UDG/UNG), heat-labile, 1 U/μL |
14466ES |
<0.1 copies/U |
|
|
UCF.METM Uracil DNA Glycosylase (UDG/UNG), 1 U/μL |
14454ES |
<0.1 copies/U |
Partial data presentation(UCF.METM Hotstart Sensitive Taq DNA Polymerase as an example)
- Residueof coli gDNA <0.005 copies/U
- coligDNA residues of different batches of UCF.METMTaq enzyme (Cat#14314ES) were detected. The result showed that E. coli gDNA residues residues of Taq DNA polymerase were well below 0.005 copies/U.
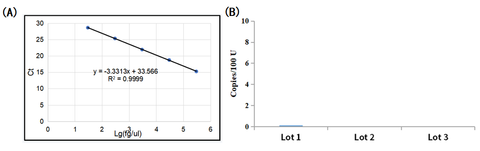
Figure 1: Detection of E.coli gDNA residue of UCF.METM Taq enzyme (Cat#14314ES)
- No residual plasmid DNA was detected
Plasmid DNA residues of UCF.METM Taq enzyme (Cat#14314ES) and Taq DNA polymerase of brand A were detected. The results showed that there was plasmid DNA residue in Taq DNA polymerase of brand A. No plasmid DNA was detected in UCF.METM Taq enzyme (Cat#14314ES).
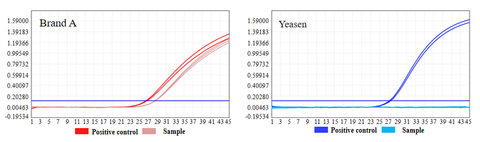
Figure 2: Results of plasmid DNA residue detection
- Eleven kinds of common background bacteria were not detected
Use the UCF.METM Taq enzyme (Cat#14314ES) coupled with primers and probes of Pseudomonas aeruginosa, Acinetobacter baumannii, Serratia marcescens, Klebsiella pneumoniae, Stenotrophomonas maltophil, Streptococcus pneumoniae, Enterococcus faecium, Staphylococcus aureus, E. coli, Enterococcus faecalis to prepare qPCR premix. NTC (No Template Control) was detected, the results showed that no residual of the above 11 common background bacteria were detected in UCF.METM Taq enzyme (Cat#14314ES).
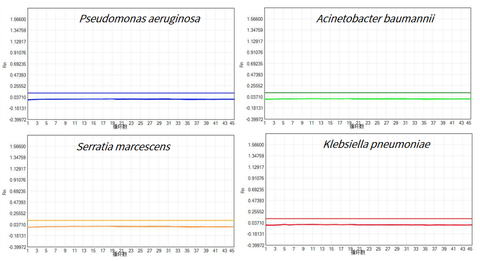
Figure 3: Test results of 11 common background bacteria (limited by space, Pseudomonas aeruginosa, Acinetobacter baumannii, Serratia marcescens and Klebsiella pneumoniae are shown as examples)
- Customer's test case presentation
The commercial Taq enzyme and YEASEN UCF.METM Taq enzyme (Cat#14314ES) were used to detect NTC together with the customer's E. coli specific primer, and the results showed that the commercial Taq enzyme peaked 5 times in 22 repeated experiments. The UCF.METM Taq enzyme (Cat#14314ES) did not peak in 22 replicates, indicating that the UCF.METM Taq enzyme (Cat#14314ES) did not detect out E. coli gDNA.
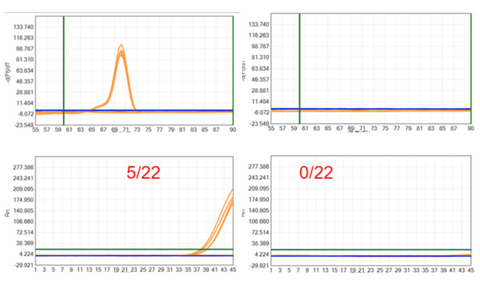
Figure 4: Detection result of E.coli gDNA residue (customer's test case)
Left: Commercially available conventional Taq enzyme; Right: UCF.METM Taq enzyme (Cat#14314ES)
Related product recommendation
|
Product Classification |
Product Name |
Catalog No. |
|
Host nucleic acid residue detection kit |
41332ES |
|
|
41331ES |
||
|
41307ES |
||
|
41308ES |
||
|
Hansenula polymorpha Host Cell DNA Residue Detection Kit |
41317ES |
|
|
E.coli Host Cell RNA Residue Detection Kit |
41318ES |
