DNase I and Their Applications in Biomedicine
Deoxyribonuclease I (DNase I) is an endonuclease, its application is not only to maintain RNA integrity but also for DNA footprint analysis, generation of random DNA libraries, reduction of stickiness in cell lysates or protein extracts, etc. In a word, DNase I can be used in almost any application that requires enzyme cleavages of DNA. The following is a detailed introduction to DNase I and its specific application.
1. What is DNase I?
2. DNase I for preparation of DNA-free RNA extraction
3. DNase I for in vitro transcription to remove template DNA
4. DNase I for rRNA removal
5. DNase I for DNA labeling
6. Other applications
7. DNase I product selection guide
1. What is DNase I?
Deoxyribonuclease I (DNase I) is a non-specific endonuclease that can digest single - or double-stranded DNA, which is present in different tissues and body fluids. It can hydrolyze phosphodiester bonds to produce mono- and oligodeoxynucleotides containing a 5'-phosphate group and a 3'-OH group. The optimal working pH range of DNase I is 7-8, its activity depends on Ca2+, and can be activated by divalent metal ions such as Mn2+, Mg2+, Zn2+, etc. In the presence of Mg2+, DNase I randomly shear any site of double-stranded DNA; in the presence of Mn2+, DNase I can shear DNA double-stranded at the same site to form a blunt end or a 1-2 nucleotide sticky ends overhangs.
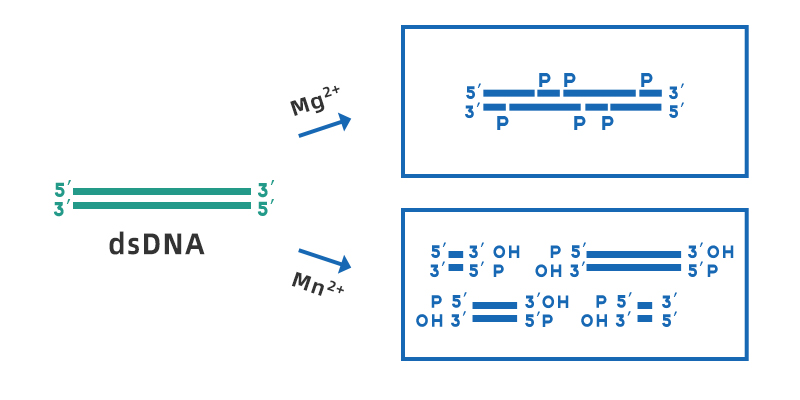
Figure 1. Schematic diagram of the cleavage of dsDNA by DNase I in the presence of Mg2+ and Mn2+.
Although DNase I cleavages are generally considered to be non-specific cleavages, DNase I is more likely to act on certain sequence fragments, such as the minor groove region, and is more prone to cleavages of purine-pyrimidine sequences. However, when DNase I act on heterogeneous dsDNA, all four bases will be cleaved, and the effect on a specific base will not be more than 3 times greater than that of other bases.
2. DNase I for preparation of DNA-free RNA extraction
In biological experiments, the first step is to prepare nucleic acid to study various functions of RNA. However, since DNA and RNA are often released together during the cell lysis process, the interference in the two cannot be avoided no matter what extraction solution is used, so specific enzymes need to be used to remove the interference. For high-quality RNA extraction, DNase I is used to removing residual DNA from the sample.
DNase I can degrade double-stranded and single-stranded DNA to oligonucleotides and single nucleotides, and the DNA in the RNA preparation product can be effectively degraded. DNase I is then inactivated by heating with a stop buffer. During the heating process, the hairpin structure of the RNA molecule can be opened, which facilitates the direct entry of the RNA into the reverse transcription process.
The quality of RNA will directly affect the experimental data to a large extent. In general, gDNA residues cannot be completely avoided during RNA extraction, therefore, it is generally recommended to treat RNA samples with DNase I to digest residual gDNA before challenging downstream applications (e.g. mRNA expression analysis, transcriptome analysis, etc.). The step of digesting gDNA can be performed during RNA extraction, after RNA extraction, or before RNA reverse transcription. According to the product positioning, the products provided by Yeasen are as follows:
Table 1: List of products related to DNA removal of RNA extraction or before reverse transcription
|
Product Positioning |
Product name |
Cat # |
|
RNA extraction |
TRIeasy™ Total RNA Extraction Reagent [inquire] |
10606ES |
|
19221ES |
||
|
MolPure™ Plant Plus RNA Kit [inquire] |
19292ES |
|
|
MolPure™ Viral DNA/RNA Kit [inquire] |
19321ES |
|
|
gDNA removal |
10325ES |
|
|
Reverse Transcription |
Hifair™Ⅲ1st Strand cDNA Synthesis SuperMix for qPCR (gDNA digester plus) |
11141ES |
|
qPCR |
11184ES |
3. DNase I for in vitro transcription to remove template DNA
In vitro transcription (IVT) mainly uses DNA as a template, plus corresponding substrates and buffers to obtain RNA through in vitro transcription. In in vitro transcription experiments, RNA polymerases such as T7, T3, and SP6 are commonly used for RNA synthesis. The synthesized RNA may have DNA residues. Eliminating the DNA residues is beneficial to the development of downstream experiments. For example, in the mRNA vaccine development stage, the removal of residues is a critical step, which can reduce the difficulty of downstream purification and increase the purity of the product. The DNA template is typically removed using Recombinant DNase I (RNase-free). According to the mRNA synthesis process, the products provided by Yeasen are as follows:
|
mRNA synthesis process |
Product name |
Cat# |
|
Template preparation |
Hieff Canace™ Plus High-Fidelity DNA Polymerase [inquire] |
10148ES |
|
10922ES |
||
|
10125ES |
||
|
FuniCut™BsaI [inquire] |
15005ES |
|
|
FuniCut™ XbaI [inquire] |
15033ES |
|
|
BspQI[inquire] [inquire] |
16215ES |
|
|
In vitro transcription |
10623ES |
|
|
10624ES |
||
|
T7 RNA polymerase (50 U/μL)[inquire] |
10618ES |
|
|
10133ES |
||
|
10620ES |
||
|
10621ES |
||
|
Remove template DNA |
10611ES |
|
|
mRNA modification |
10614ES |
|
|
10612ES |
||
|
10132ES |
||
|
10619ES |
||
|
mRNA purification |
12602ES |
4. DNase I for rRNA removal
In vivo, rRNA is highly abundant and very conservative, which is of little significance in obtaining biological information, so rRNA is often removed first in RNA library construction and sequencing. At present, the removal method of rRNA is mainly RNase H digestion. The main steps of enzymatic-based rRNA depletion are shown in figure 2: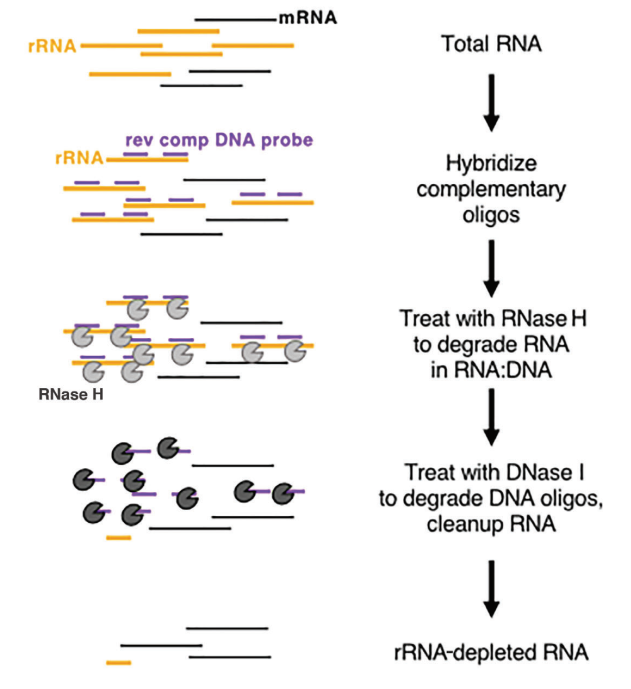
Figure 2: Schematic diagram of the principle of enzymatic-based rRNA depletion (Baldwin, A. et al. 2021, Current Protocols)
First, extract total RNA, then hybridize single-stranded DNA probe with rRNA, design and synthesize rRNA-specific single-stranded DNA probe, then use RNase H to degrade the hybridized rRNA, and use DNase I to degrade the DNA probe. At last, leaving the non-rRNA RNA template. The products related to rRNA removal provided by Yeasen are as follows:
|
mRNA synthesis process |
Product name |
Cat# |
|
Human/Mouse/Rat rRNA Depletion |
Hieff NGS™ MaxUp rRNA Depletion Kit (Human/Mouse/Rat) MaxUp [inquire] |
12253ES |
|
Plant rRNA Depletion |
12254ES |
|
|
Removal of ribosomal RNA and 45S ITS/ETS regions from human total RNA |
12257ES |
|
|
Degradation of rRNA |
12906ES |
|
|
DNA probe degradation |
10325ES |
5.DNase I for DNA labeling
The nick translation is one of the laboratory's most commonly used deoxyribonucleic acid probe labeling methods. This method utilizes various enzymatic activities of DNA polymerase I to incorporate labeled deoxyribonucleoside triphosphates into newly synthesized DNA chains. Thus, uniformly labeled DNA probes for high specific activity are synthesized. The characteristics of nick translation are fast, simple, deliberate, high specificity, and uniformly labeled probes, which are suitable for longer double-stranded DNA.The method is realized by the combined action of DNase I and E. coli DNA Polymerase I. The main steps of DNA labeling by nick translation are shown in figure 3:
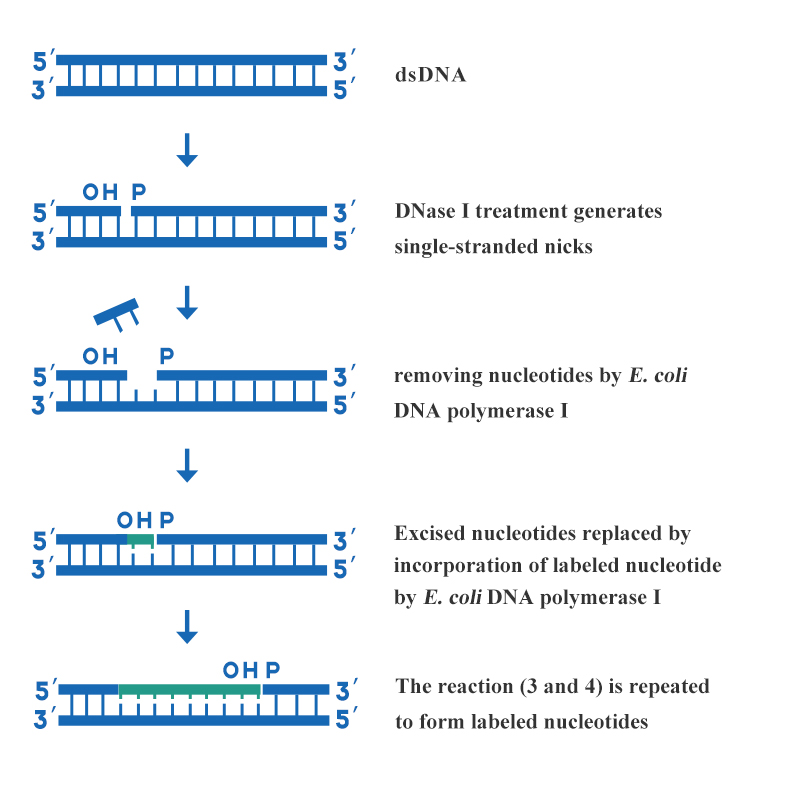
Figure 3: Schematic diagram of DNA labeling by nick translation
A suitable concentration of DNase I is used to creating several single-stranded gaps on each strand of the double-stranded DNA to be labeled, and the 3' hydroxyl terminus is formed at the gap. Use the 5'→3' exonuclease activity of E. coli DNA Polymerase I to cut a nucleotide from the 5' end of the nick, and at the same time the 5'→3' polymerase activity of E. coli DNA Polymerase I introduce a nucleotide labeled with the 3' end of the gap to repair the gap. As the gap moves along the DNA strand, the labeled nucleotides are incorporated into the newly synthesized strand. The products related to DNA labeling provided by Yeasen are as follows:
|
Product positioning |
Product name |
Cat# |
|
Ordinary |
Deoxyribonuclease I (DNase I) from bovine pancreas [inquire] |
10607ES/10608ES |
|
RNase free |
10325ES |
|
|
E.coli source |
12903ES |
6. Other applications
The above are several commonly used applications. More applications of DNase I include the following, such as DNase I footprinting assay and DNase I hypersensitive sites. DNase I footprinting assay is a detection method that can accurately identify the binding sites of DNA-binding proteins on DNA. When a protein binds to a DNA fragment, it can protect the binding site from being damaged by DNase I and the DNA fragments will be left behind after enzyme digestion ("footprint"), and its sequence can be determined. In the gel image, there are no bands where the DNA binds to the protein. To read more click on the link.DNase I hypersensitive sites refer to cleavages at a small number of specific sites when chromatin is treated with low DNase I and these specific sites are called DNase I hypersensitive sites. The principle is that when a gene is in a transcriptionally active state, the chromatin containing the gene is significantly more sensitive to DNase degradation than the inactive region. To read more click on the link.
7. DNase I product selection guide
Yeasen Biotechnology (Shanghai) Co., Ltd., founded in 2014, is a high-tech enterprise engaged in the R &D and production of tool enzyme raw materials and antigen antibodies. Its products include molecular diagnostic enzymes, proteins, and antibodies used in pharmaceuticals, food safety testing, breeding, justice, and other industries. We are committed to providing customers in the field of life sciences with high-quality products and services. The product purchase guidelines for DNase I are as follows:|
Product name(Cat#) |
Product positioning |
Recommended applications |
|
DNase I from bovine pancreas (CAT#10607,10608)[inquire] |
RNase removed, not detected |
Mainly used in protein research: DNA removal from protein preparations. |
|
Recombinant DNase I (RNase-free)(CAT#10325) |
RNase-Free, for research |
Ideal for a variety of applications: DNA removal from RNA and protein preparations such as RNase-sensitive cDNA libraries or sample preparation for RT-PCR experiments. |
|
RNase-Free, GMP pharmaceutical grade. |
Ideal for a variety of applications: DNA removal from RNA and protein preparations such as RNase-sensitive cDNA libraries or sample preparation for RT-PCR experiments. |
Regarding reading:
GMP-grade reagents for mRNA in vitro synthesis
The principles of DNase I footprinting and its biomedical applications
References
1. Baldwin A, Morris A R, Mukherjee N. An Easy, Cost-Effective, and Scalable Method to Deplete Human Ribosomal RNA for RNA-seq[J]. Current Protocols, 2021.
2. Song C, Zhang S, Huang H. Choosing a suitable method for the identification of replication origins in microbial genomes[J]. Frontiers in Microbiology, 2015, 6:1049.
