High-quality isothermal amplification raw materials ——Making RT -LAMP more sensitive and faster!
Isothermal amplification technology can achieve the purpose of amplifying specific nucleic acid sequences under constant temperature conditions. Due to the characteristics of no special equipment, short amplification time, and high sensitivity, it has shown good application prospects in on-site detection and point-of-care diagnosis and is very suitable for the development of molecular diagnostic tools.
In the context of the novel coronavirus pandemic with increasing strains and infectivity, Isothermal amplification technology can detect the novel coronavirus rapidly with simple instruments, helping to solve the problem that some COVID-19 patients can't be quickly confirmed due to the long detection time and high requirements for detection equipment and reagents of traditional PCR techniques. So what is the principle of RT-LAMP, one of the isothermal amplification techniques? What are the high-quality raw materials that Yeasen can provide?
1. What is the principle of RT-LAMP?
2. Primer design for RT-LAMP
3. What raw materials can Yeasen provide?
4. Product Selection Guide
1. What is the principle of RT-LAMP?
In 2000, Japanese scholars Notomi and others established a new nucleic acid amplification technology called loop-mediated isothermal amplification (LAMP). LAMP uses 4 specific primers designed for 6 regions of the target gene, and uses strand displacement DNA polymerase to amplify 109 copies of the target sequence in tens of minutes under isothermal conditions (60~65 ℃). The results were judged by agarose gel electrophoresis, and the positive results showed ladder-shaped bands. LAMP has the characteristics of strong specificity, high sensitivity, fast and simple operation, and low cost. With the continuous improvement and perfection of this technology, it is currently used in the detection of various pathogenic microorganisms.
RT-LAMP is a method in which reverse transcriptase is added to the LAMP amplification system, which can realize the direct detection of viral RNA. The amplification efficiency of LAMP is extremely high, so a large amount of nucleic acid can be amplified with only a small amount of cDNA. The time of reverse transcription in the nucleic acid amplification process is saved, and the detection speed of RNA is accelerated.
DNA is in a state of dynamic equilibrium at about 65℃. Under the action of strand displacement DNA polymerase, starting from the 3' end of the F2 segment of the FIP primer, it is paired with the complementary sequence of the template DNA to initiate strand displacement DNA synthesis. The F3 primer is complementary to F3c at the front end of F2c and takes the 3' end as the starting point to synthesize its DNA while replacing the DNA strand synthesized by the leading FIP primer by the action of a strand displacement DNA polymerase. Extend forward. The DNA strand synthesized by the final F3 primer forms a double strand with one template DNA strand. The DNA strand synthesized by the FIP primer is replaced by the F3 primer strand to generate a single strand. This single strand has complementary F1c and F1 segments at the 5' end, so self-base pairing is performed to form a circular structure. And the BIP primer is hybridized and combined with the single strand, and the 3' end of the BIP primer is used as the starting point to synthesize a complementary strand, and the structure is opened in the process. Then, primers similar to F3 and B3 are inserted from the BIP primer, base complementary pairing is performed, and a new complementary strand is synthesized from the 3' end as the starting point. There are complementary sequences at both ends of the replaced single-stranded DNA, and self-base pairing occurs to form a circular structure, so the entire chain presents a dumbbell-like structure. This structure is the starting structure of the amplification cycle of the RT-LAMP method.
In the dumbbell-shaped structure, the DNA extension is carried out using the F1 segment at the 3' end as a starting point and using itself as a template. And the FIP primer F2 hybridizes with the single-stranded F2c on the loop to initiate a new round of strand displacement reaction. The double-stranded nucleic acid synthesized from the F1 segment is dissociated, and similarly, a circular structure is formed on the nucleic acid. There is a single-stranded form of B2c on the circular structure, and B2 on the BIP primer hybridizes with it to initiate a new round of amplification, and a circular structure is formed through the same process. According to this process, complementary sequences on the same strand cycle through pairing, strand extension, and finally form structures of different sizes.
2. Primer design for RT-LAMP
Primer design is the key to the successful amplification and detection of RT-LAMP. RT-LAMP primers include two outer primers (F3 and B3) and two inner primers (FIP and BIP). F3 primer: The upstream outer primer, consisting of the F3 region, is complementary to the F3c region of the target gene. FIP primer: upstream internal primer, consisting of the F2 region, the F2c region at the 3' end of the target gene in the F2 region is complementary to the F1c region at the 5' end of the target gene. BIP primer: downstream internal primer, composed of B2 region, B2 region is complementary to the B2c region at the 3' end of the target gene, and has the same sequence as the B1c region at the 5' end of the target gene.
Similar to PCR, primer design principles should pay attention to factors such as base composition, GC content, and substructure. In addition, the following points should be noted. The 5' end portions of the inner primers FIP and BIP, ie, F1c and B1c, are generally 8-50 bp in length. The length is preferably 15 to 25 bp, and the Tm value is greater than the Tm values of F2 and B2 at the 3' end portion. The 3' end portions of the inner primers FIP and BIP, ie, F2 and B2, are generally 8-50 bp in length. The length is preferably 15-25bp, and the Tm value is consistent with the optimum temperature of the Bst DNA polymerase selected in the experiment. The lengths of the outer primers F3 and B3 are generally 8-50 bp. The length is preferably 15-25 bp, and its Tm value is smaller than that of F2 and B2. When designing primers, the loop structure and the size of the target sequence should be considered. When the number of bases in the loop is greater than 40bp and the size of the target sequence is 130-200bp, the amplification efficiency is the highest.
3. What raw materials can Yeasen provide?
3.1 [New upgrade] Yeasen Bst Plus DNA Polymerase
The newly upgraded Hieff™ Bst Plus DNA Polymerase (Cat#14402ES, 14403ES) is obtained by expressing and purifying the DNA polymerase gene of Bacillus stearothermophilus sp lacking the 5′→3′ exonuclease domain in E.coli. The enzyme strand displacement ability is strong, and has the advantages of high sensitivity, high amplification efficiency, and high dUTP tolerance, and can be widely used in the real-time detection of pathogens based on isothermal amplification technology.
3.1.1 Fast and high sensitivity
Yeasen Hieff™ Bst Plus DNA Polymerase has high sensitivity and can detect the target gene as low as 5 copies. The amount of template is at the femtogram (fg) level, and the amplification rate is faster than competing products.
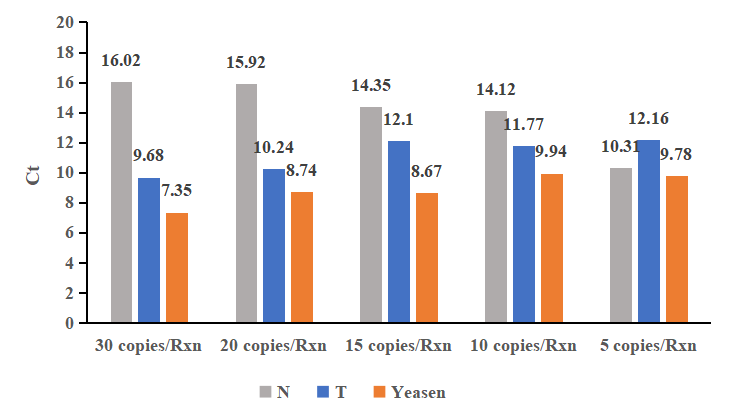
Figure 1. RT-LAMP reaction was carried out with Yeasen Hieff™ Bst Plus DNA Polymerase (Orange) and Bst enzymes of competitor N (Gray) and competitor T (Blue) to amplify SARS-CoV-2. The results show that Yeasen Hieff™ Bst Plus DNA Polymerase can always reach the threshold faster than competing products, and the amplification rate is faster.
4. Product Selection Guide
The products provided by Yeasen are as follow:
Table 1. Product information
|
Product Positioning |
Product name |
Cat# |
|
Highly sensitive Bst enzyme |
14402ES |
|
|
Hieff™ Bst Plus DNA Polymerase (2000 U/μL) (Inquire) |
14403ES |
|
|
Fluorescence Dye Method RT-Lamp Display Kit |
13762ES |
|
|
Reverse transcriptase suitable for RT-Lamp |
Hifair™ Ⅲ Reverse Transcriptase (Inquire) |
11111ES |
|
Hifair™ Ⅲ Reverse Transcriptase, Glycerol-free (Inquire) |
11297ES |
|
|
Murine RNase Inhibitor |
10603ES |
|
|
10703ES |
||
|
Heat-labile UDG |
Uracil DNA Glycosylase (UDG/UNG), heat-labile, 1 U/μL (Inquire) |
10303ES |
|
High-purity dUTP |
10128ES |
Regarding reading:
